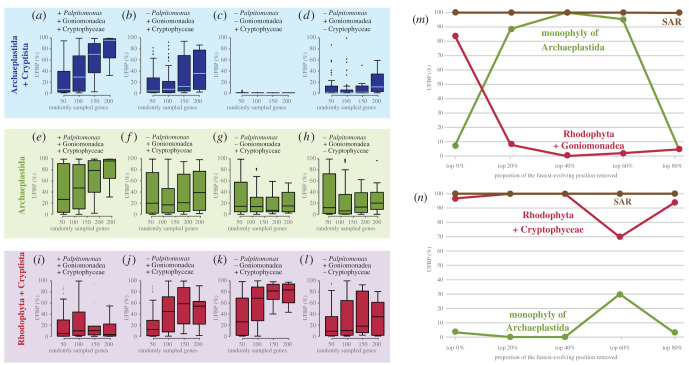
Figure 3.
Analyses assessing the phylogenetic affinity of Rhodophyta to Cryptophyceae and/or Goniomonadea. (a–l) Analyses of the alignments generated by random gene sampling (RGS). We excluded both Rhodelphis spp. and Microheliella maris from the ‘rs50g,’ ‘rs100g,’ ‘rs150g’ and ‘rs200g’ alignments, which were generated from the Diaph alignments (see Methods for the detail) and then subjected to the ultrafast bootstrap analyses using IQ-TREE 1.6.12. The ultrafast support values (UFBPs) for the sister relationship between Archaeplastida and Cryptista, the monophyly of Archaeplastida, and the union of Rhodophyta and Cryptista are presented as box-and-whisker plots (a), (e) and (i), respectively. The ultrafast bootstrap analyses on the rs50g, rs100g, rs150g and rs200g alignments were repeated after further exclusion of Palpitomonas bilix (b, f and j), P. bilix and Goniomonadea (c, g and k), and P. bilix and Cryptophyceae (d, h and l). The UFBPs shown in the plots described above are summarized in electronic supplementary material, table S4. (m,n) Analyses of the alignments processed by fast-evolving position removal (FPR). We modified the Diaph alignment in two ways, (i) the exclusion of Rhodelphis spp., M. maris, P. bilix, and Cryptophyceae and (ii) that of Rhodelphis spp., M. maris, P. bilix and Goniomonadea. The two modified Diaph alignments were processed by FPR and further subjected to ultrafast bootstrap analyses. We plotted the UFBPs for the monophyly of the SAR clade (brown), those for the monophyly of Archaeplastida (green), and those for the uniting of Rhodophyta and Goniomonadea/Cryptophyceae (red).