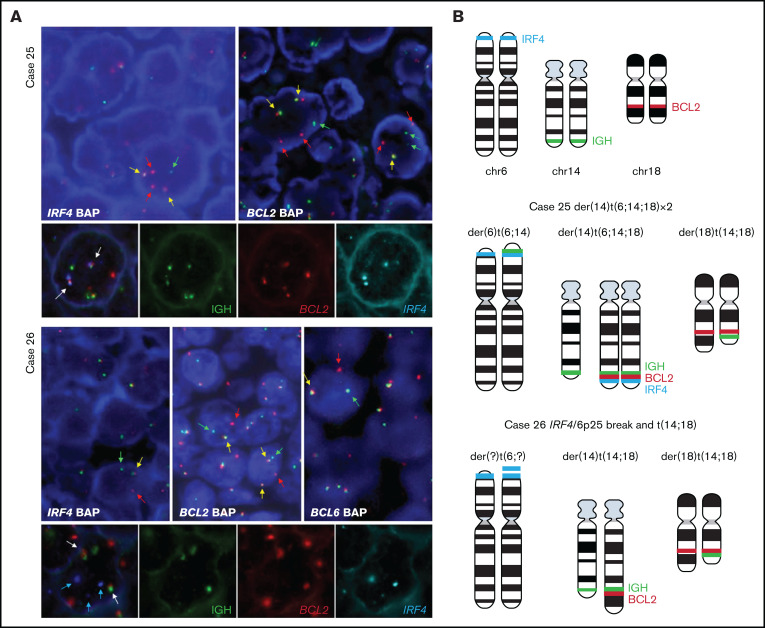
Figure 2.
FISH analysis demonstrates complex rearrangement with IRF4 and BCL2/BCL6 genes. (A) FISH analyses using IRF4, BCL2, and BCL6 break-apart probes and triple-color FISH for IGH, BCL2, and IRF4 loci in the 2 cases with IRF4 rearrangement together with BCL2 in case 25 and BCL2/BCL6 rearrangement in case 26. These hybridizations were performed using t(14;18) dual-color fusion (Metasystems) and BAC clones spanning IRF4 locus (BAC clones 5' RP3-416J7 and 3' RP5-1077H22 and RP5-856G1) labeled in spectrum aqua. Red signals correspond to BCL2, green signals correspond to IGH, and aqua/blue signals represent IRF4. Case 25. FISH demonstrates IRF4 and BCL2 breaks (upper panel) with 1 or 2 colocalized signals (yellow arrows) and 1 or 2 split signals (green and red arrows) consistent with BCL2 and IRF4 gene rearrangement. The triple rearrangement (IGH-IRF4-BCL2) (lower panel) is demonstrated by colocalization of the 3 colors (white arrows). Case 26. FISH analysis for IRF4, BCL2, and BCL6 with break-apart probes demonstrates 1 colocalized signal (yellow arrow) and 1 split signal (green and red arrows), indicating a triple rearrangement. FISH analyses of concomitant IGH, BCL2 and IRF4 rearrangements (lower panel) show 2 colocalized signals (white arrow) of the derivative chromosomes 14 and 18 resulting in t(14;18) translocation. IRF4 locus (blue arrows) did not colocalize with either the IGH locus or the t(14;18) translocation, indicating that this is an independent rearrangement with unknown partner. (B) Schematic representation of IRF4 (aqua blue), IGH (green), and BCL2 (red) breaks/rearrangements in cases 25 and 26.