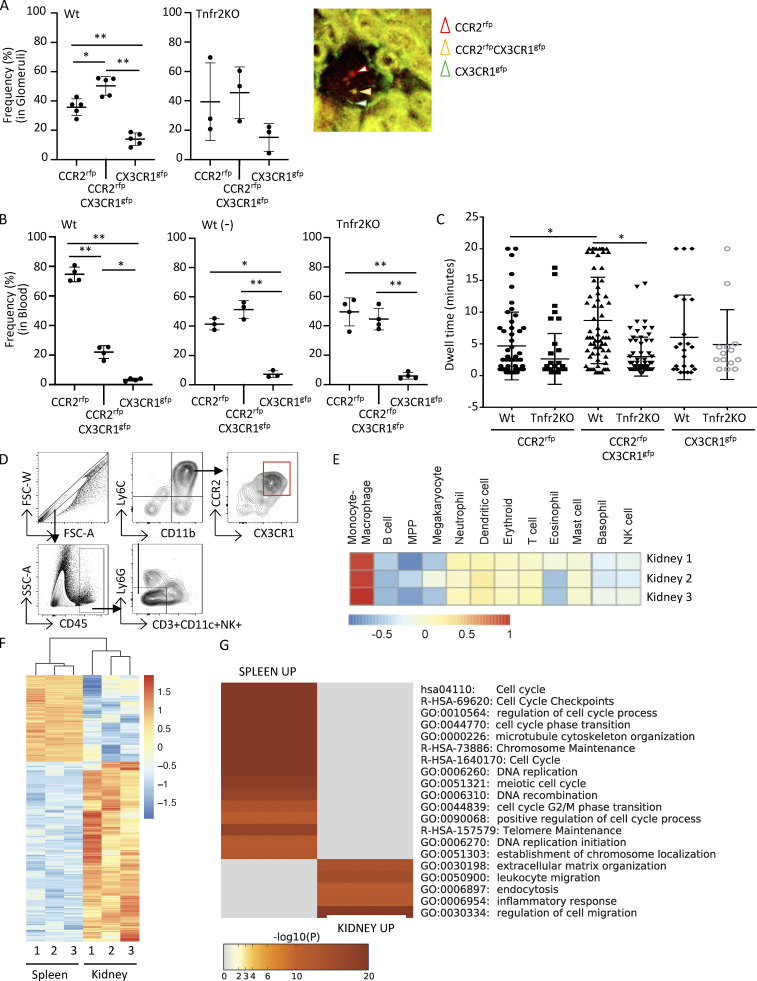
Figure 2.
Renal accumulated Ly6C+CCR2+CX3CR1+ monocytes exhibit an increase in intravascular dwell time within glomerular capillaries and have a proinflammatory transcriptional profile. (A–C) Radiation chimeric Wt and Tnfr2 KO recipient mice reconstituted with Ccr2rfp/+Cx3cr1gfp/+ (red signal, CCR2-rfp; green signal, CX3CR1-gfp) reporter bone marrow were analyzed on day 10 after NTN induction. (A) Two-photon IVM of glomeruli was undertaken, and images were acquired every 30 s for 20 min. The frequency of monocyte subpopulations among recruited cells, defined as a cell adherent for two frames (60 s) and a representative image of a glomerulus with cells positive for RFP (CCR2rfp), GFP (CX3CR1gfp), or both fluorophores (CCR2rfpCX3CR1gfp), is shown. Three to five mice per group and three to four glomeruli were imaged per mouse across six independent experiments. (B) Blood taken before (−) and after induction of NTN in Wt chimeras and after NTN in Tnfr2 KO chimeras were analyzed by flow cytometry. (C) The dwell time of individual monocytes within glomeruli analyzed in A is given. One-way ANOVA (Tukey’s multiple comparison test) was performed. *, P < 0.005; **, P < 0.0001. (D) FACS sorting strategy for isolating Ly6C+ monocytes expressing both CCR2 and CX3CR1 using lineage-negative markers to exclude T cells, dendritic cells, and NK cells and gating remaining cells for CD11b and Ly6C and then CCR2 and CX3CR1. CCR2 and CX3CR1 DP cells were collected. FSC, forward scatter; SSC, side scatter. (E) Haemopedia cell type fingerprint heatmap. Shown are the mean row-normalized log2 CPM values for the indicated gene sets, clustered by gene set for the renal DP population. (F) Heatmap of DEGs in renal DP cells vs. spleen DP cells. Shown are row-normalized log2 CPM values. (G) Top 20 GO terms enriched in the DEGs of E. Data are from three biological replicates, with each replicate containing sorted kidney and spleen/bone marrow cells from 10 mice.