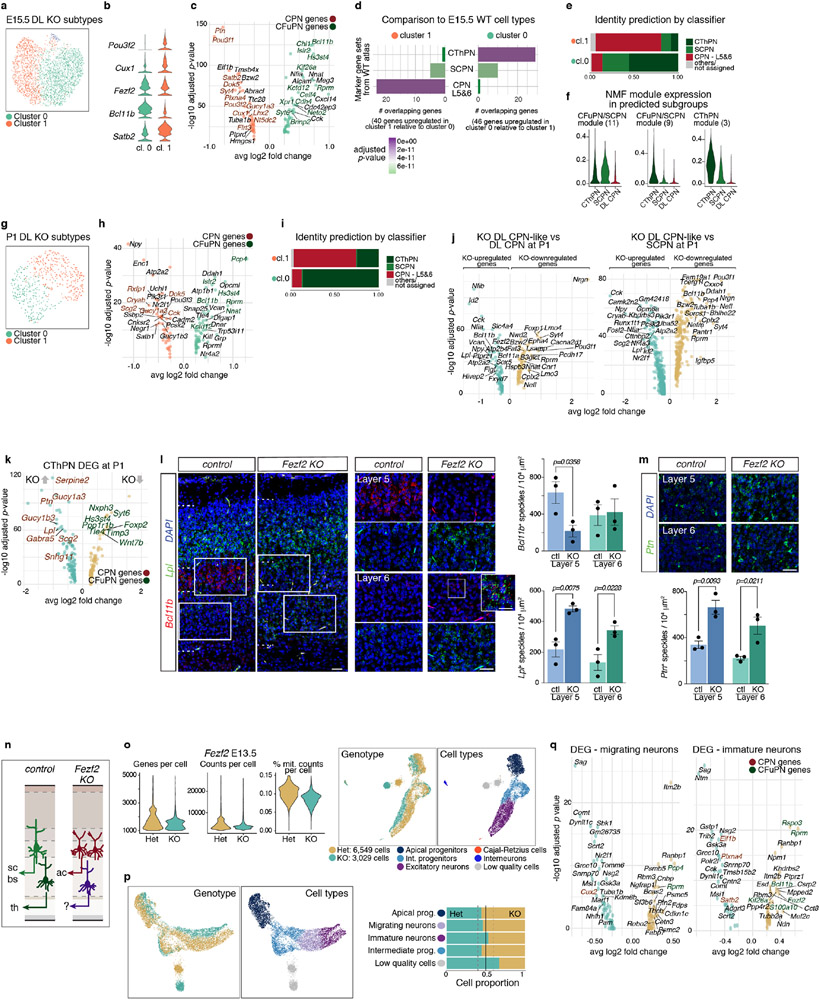
Extended Data Figure 12 (related to Figure 5). CFuPN acquire CThPN-like and layers 5&6 CPN-like identities in the absence of Fezf2.
a-f Two subtypes of deep-layers KO cells were Identified at E15.5. Sub-clustering of deep-layers KO-exclusive cells alone at E15.5 (a) shows a Satb2LOW, Bcl11bHIGH cluster (cluster 0), and a Satb2HIGH cluster expressing also CPN markers Cux1 and Pou3f2 (cluster 1), as indicated in the violin plots (b). Differential expression analysis between both subtypes indicates enrichment of CFuPN genes in cluster 0 and CPN genes in cluster 1 (c). d Comparison to neurons in E15.5 wild-type data showing overlap between differentially expressed genes and markers from E15.5 neuronal subtypes. Bars indicate number of overlapping genes and are colored by the adjusted p-value calculated by hypergeometric test for significant enrichment. e Classification of cells from both E15.5 KO-specific clusters according to random forest classifier shows good agreement between both annotations. f NMF module expression (as in Fig. 5b) in the KO-specific cells, grouped according to the cell type assigned by the random forest classifier. g-i Sub-clustering (g) and differential expression analysis (h) of deep-layers KO-exclusive cells alone at P1 reveals two subpopulations that correspond to CThPN-like and layers 5&6 CPN-like populations. i Classification of cells from both P1 KO-specific clusters according to random forest classifier shows good agreement between both annotations.
j-k Differential expression analysis of the aberrant layer 5&6 CPN-like cells from the KO-exclusive populations at P1 compared to layers 5&6 CPN (j) or SCPN (k) populations in the control.
l-m In situ hybridization against Bcl11b and Lpl (a) or Ptn (b), in P1 control (wild type) and Fezf2 KO coronal sections, showing higher levels of expression of Lpl and Ptn on layers 5 and 6 and reduced Bcl11b in layer 5 (insets to the right correspond to boxes in left panels). Note cells expressing both Bcl11b and Lpl in magnification from layer 6, reflecting an aberrant CThPN identity. Number of positive speckles per 104 μm2. Quantification was calculated with a modified pipeline from CellProfiler from an area of ~200 by 150 μm or ~200 by 100 μm centered in layers 6 or 5, respectively. Data correspond to mean±sem, from n = 3 mice, > 3 sections each. Unpaired t test, exact p-values indicated. Scale bars are 30 μm, except in higher magnification in l, 15 μm.
n Violin plots of number of genes (left) and mRNA molecules (counts; middle), and percentage of mitochondrial counts (right) per cell in control and KO Fezf2 E13.5 single cell transcriptomes, and UMAP visualizations of combined control and KO complete data sets, colored by genotype or assigned cell type. o Dorsally-derived cells in Fezf2 control and KO E13.5 scRNA-seq, visualized via UMAP and colored by genotype (left) or cell types (right). Proportion of cells in each cell type, according to their genotype. p Differential expression analysis between control and KO migrating or immature neurons shows upregulation of a subset of CPN marker genes and downregulation of CFuPN-specific genes.