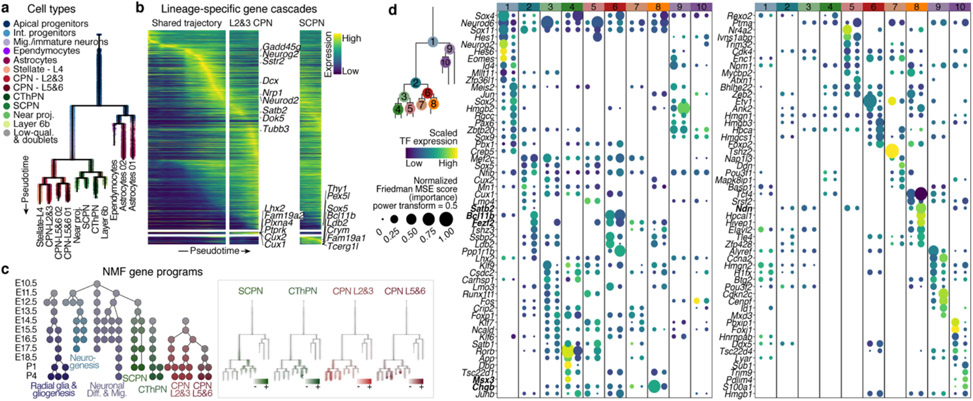
Figure 3. Molecular developmental trajectories of neocortical cell types.
a URD branching tree. Root is E10.5 earliest progenitors, tips are P4 terminal cell types. Cells colored by their identity.
b Smoothened heatmap of gene cascades for layers 2&3 CPN and SCPN. Gene expression in each row is scaled to maximum observed expression, and smoothened. Genes are ordered based on their onset and peak expression timings. Some marker genes are labeled. The cascade is divided into three segments: shared trajectory, layer 2&3 CPN-specific and SCPN-specific trajectories. Full cascades in Extended Data Fig. 7 and Supplementary Information Table 3.
c Gene programs of connected modules found by NMF. Left: circular nodes represent modules aligned by the age they were computed from. Right: scaled expression of lineage specific modules on the branching tree.
d Genes predicted to be involved in cell type divergence. Top 10 transcription factors per branch, ranked by their feature importance score for ability to distinguish between branches (Friedman MSE score, 0.5 power transformed, dot size), and their average expression in the corresponding cells (row-scaled, color). Color bar at the top indicates branch-points.