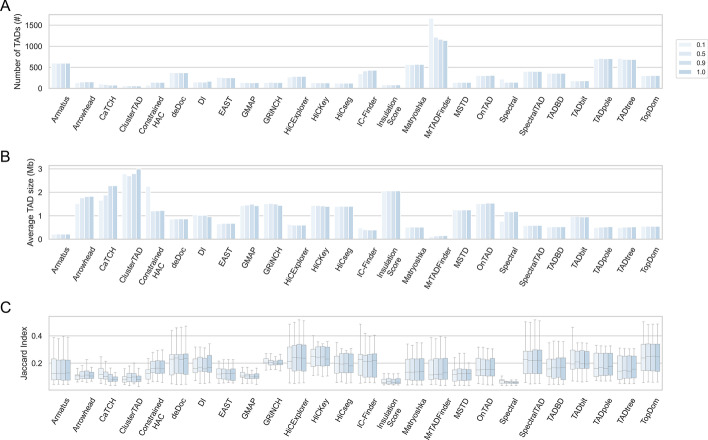
Fig. 3.
TAD inference performance via TAD callers across a number of sequencing depths at 5kb resolution using HiCNorm normalized Micro-C interaction dataset over human embryonic stem cell line. A Across multiple ratio of downsampled sequenced reads, the number of TADs inferred by each caller in all chromosomes. B The mean TAD size inferred by considered TAD callers across different ratio of downsampled sequenced reads over all chromosomes. C The comparison of TAD boundaries inferred by one TAD caller with the others across a number of sequencing depths over all chromosomes in terms of Jaccard Index similarity metric. We do not report CHDF results as CHDF has inferred each bin in the interaction matrix as an independent TAD after downsampling the dataset