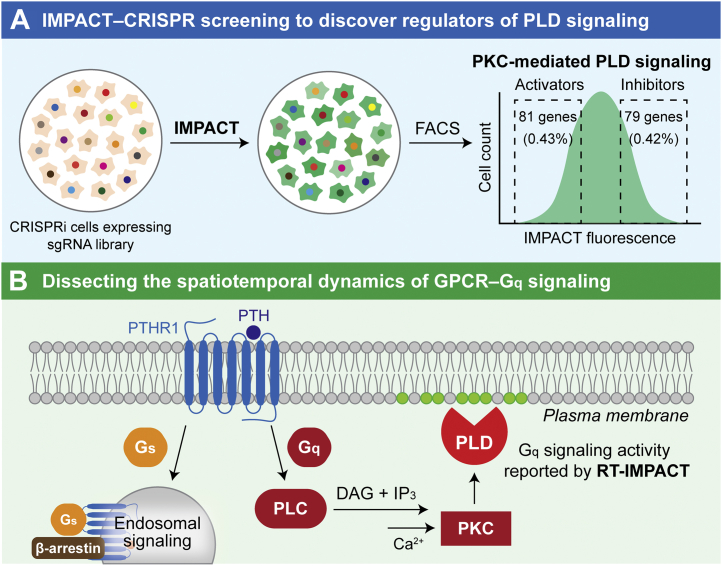
Figure 4.
Applications of IMPACT and RT-IMPACT to understand PLD-mediated cell signaling.A, IMPACT-enabled CRISPR screens have the potential to reveal new regulatory circuits controlling PLD signaling. CRISPRi cells expressing a dead Cas9–KRAB fusion and one member of a genome-wide sgRNA library are labeled with IMPACT, followed by FACS-based enrichment of the IMPACT-high and IMPACT-low cell populations. Cells with a higher extent of IMPACT labeling contain sgRNAs targeting putative PLD-inhibitory genes, and those with lower levels of IMPACT labeling contain sgRNAs targeting putative PLD-activating genes. Identities of such putative PLD inhibitors and activators are determined by next-generation sequencing of sgRNAs in enriched populations relative to each other or unsorted cells. B, RT-IMPACT enables visualization and quantification of the subcellular locations and duration of PLD signaling elicited by different stimuli. PTH-induced activation of PTHR1 leads to two separate downstream signaling pathways, Gs and Gq, that are spatiotemporally controlled. RT-IMPACT can be used to dissect the two signaling events by selectively revealing PLD activation that occurs downstream of Gq signaling. FACS, fluorescence-activated cell sorting; GPCR, G protein-coupled receptor; IMPACT, Imaging PLD Activity with Clickable Alcohols via Transphosphatidylation; PLC, phospholipase C; PLD, phospholipase D; PTHR1, parathyroid hormone receptor.