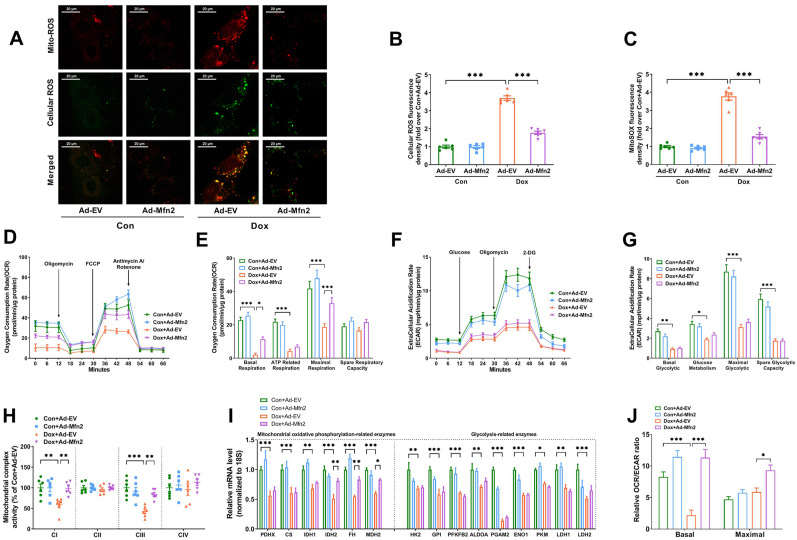
Fig. 3.
Mfn2 overexpression inhibited mitochondrial ROS and enhanced mitochondrial oxidative metabolism in the Dox-treated cardiomyocytes. (A) Representative images of MitoSOX-stained mitochondrial ROS (red fluorescence) and DCFH-DA-stained cellular ROS production (green fluorescence). Original magnification × 600. (B) Quantitative data of relative whole-cell ROS fluorescence density expressed as a fold change compared with Con + Ad-EV. (C) Quantitative data of relative mitochondrial ROS fluorescence density expressed as a fold change compared with Con + Ad-EV. (D-E) Oxygen consumption rate (OCR) and quantitative data of OCR. (F-G) Extracellular acidification rate (ECAR) and quantitative data of ECAR. (H) Quantitative data of mitochondrial complex I to IV (CI to CIV) activity expressed as a fold change compared with Con + Ad-EV. n = 6 independent experiments per group in Figure A–C and Figure H. (I) Quantitative mRNA expression of representative metabolic enzymes involved in mitochondrial oxidative phosphorylation and glycolysis. PDHX, pyruvate dehydrogenase complex component X; CS, citrate synthase; IDH, isocitrate dehydrogenase; FH, fumarate hydratase; MDH, malate dehydrogenase; HK, hexokinase; GPI, glucose-6-phosphate isomerase; PFKFB2, 6-phosphofructo-2-kinase; ALDOA, fructose bisphosphate aldolase A; PGAM2, phosphoglycerate mutase 2; ENO1, enolase 1; PKM, Pyruvate kinase M; LDH, lactate dehydrogenase. (J) The ratio of oxygen consumption rate (OCR) relative to extracellular acidification rate (ECAR). n = 3 independent experiments per group in Figure D–G and FigureI-J. One-way ANOVA with Turkey's multiple comparison test was used. *P < 0.05, **P < 0.01, ***P < 0.001. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)