FIG 2.
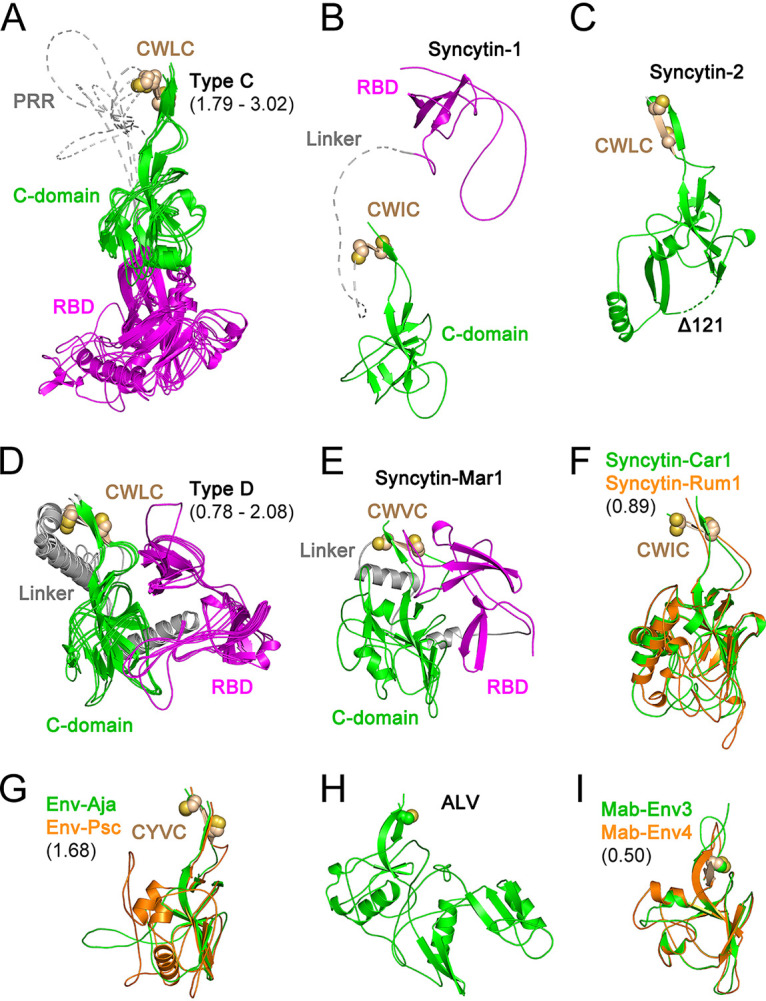
Orthoretroviral SU structural models. Shown are models of the SUs of type C gammaretroviruses (A), syncytin-1 (B), syncytin-2 (C), type D gammaretroviruses (D), syncytin-Mar1 (E), syncytin-Car1 and Rum1 (F), Env-Aja and Env-Psc (G), ALV (H), and Mab-Env3 and Mab-Env4 (I). Panels A, D, F, G, and I show superpositions of structurally similar models, with minimum and maximum root mean square deviations of aligned models in angstroms shown in parentheses. The models in panels A, B, D, and E show the RBD in magenta and the linker regions joining the RBD to the C-domain in gray. The models in panels F, G, and I are shown in colors as indicated in each panel. The disordered PRR and linker regions are shown as dotted lines in panels A and B. The conserved cysteine residues in the gammaretroviral CWLC consensus motifs in panels A to G or preceding the first β-strand in the alpharetrovirus SU models in panels H and I are shown as spheres. The deletion in the syncytin-2 SU model (Δ122) is shown as a dotted line.
