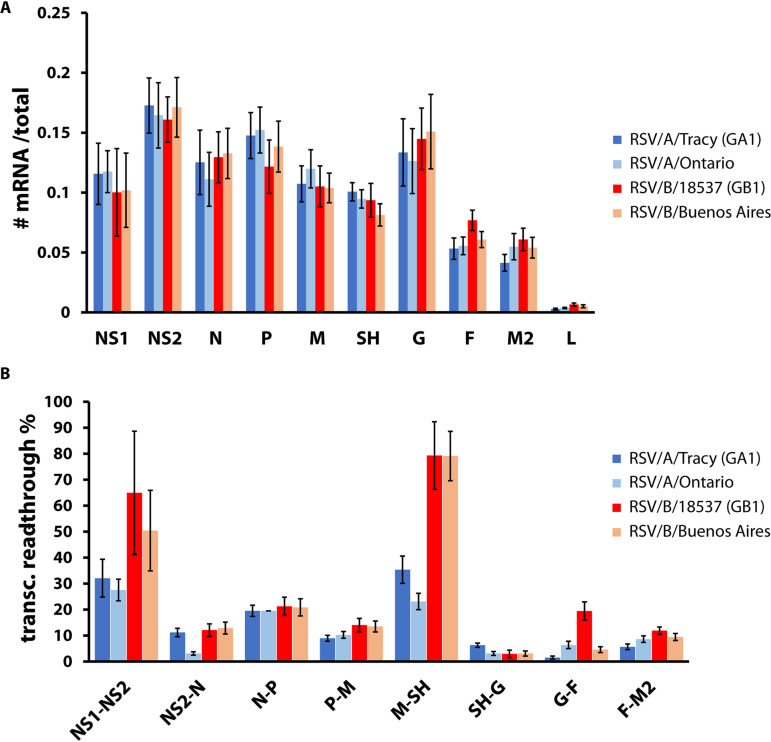
FIG 2.
RSV gene expression. HEp-2 cells were infected with RSV (RSV/A/Tracy [GA1], RSV/A/Ontario, RSV/B/18537 [GB1], RSV/B/Buenos Aires) at a multiplicity of infection (MOI) of 0.01. Samples were collected at 24 and 48 h postinoculation (hpi). RNA was isolated from cell lysates and prepared for and subjected to high-throughput short-read sequencing. (A) Relative mRNA levels are comparable across the four RSV strains tested. For each time point and each of 10 RSV genes, the average read depth across the gene’s coding sequence was divided by the total number of reads mapping to all 10 viral coding sequences. Data shown are averages ± SD of results for 24 and 48 hpi. (B) Transcriptional readthrough at particular gene junctions varied between RSV subgroups and strains. For each time point and each of eight RSV gene junctions, the average read depth across the gene junction was divided by the average read depth across the nearest upstream coding sequence. Data shown are averages ± SD of results for 24 and 48 hpi.