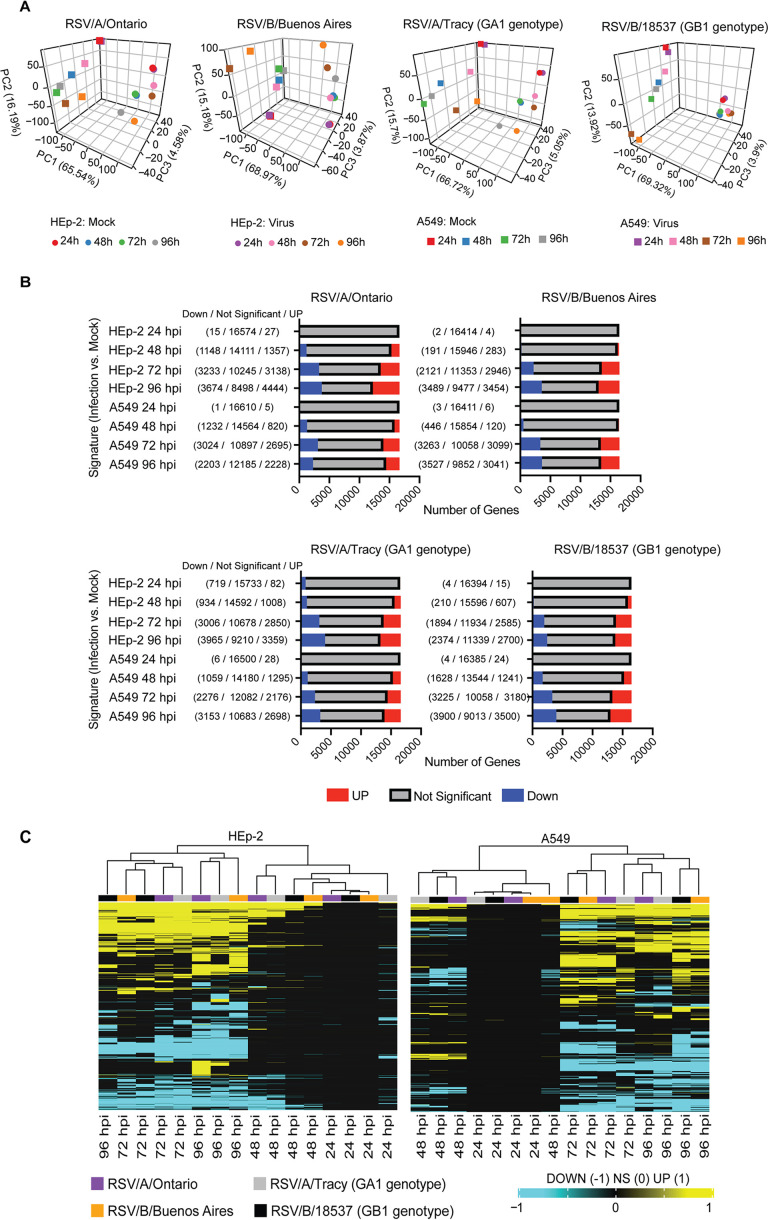
FIG 6.
RNA sequencing analysis of RSV infections of HEp-2 and A549 cells. (A) Principal-component analysis of RSV/A/Ontario- (ON), RSV/B/Buenos Aires- (BA), RSV/A/Tracy- (GA1), and RSV/B/18537- (GB1) infected HEp-2 and A549 cells demonstrating the variability observed in the samples (HEp-2 versus A549 and infected versus mock-infected controls). (B) Total number of significant genes identified for the RSV/A/Ontario-, RSV/B/Buenos Aires-, RSV/A/Tracy-, and RSV/B/18537-infected group in HEp-2 and A549 cells (adjusted P < 0.05). (C) Samples were hierarchically clustered based on the union of differentially expressed genes across all comparisons (false discovery rate [FDR] <0.05 and fold change exceeding 1.5×). Genes were converted to 1 for upregulated; −1 for downregulated; and 0 for no significant upregulation or downregulation in a particular signature. Samples were clustered using the Euclidean distance.