Abstract
Objectives:
Hereditary hearing loss exhibits high degrees of genetic and clinical heterogeneity. To elucidate the population-specific and age-related genetic and clinical spectra of hereditary hearing loss, we investigated the sequencing data of causally associated hearing loss genes in a large cohort of hearing-impaired probands with a balanced age distribution from a single center in Southwest Germany.
Design:
Genetic testing was applied to 305 hearing-impaired probands/families with a suspected genetic hearing loss etiology and a balanced age distribution over a period of 8 years (2011–2018). These individuals were representative of the regional population according to age and sex distributions. The genetic testing workflow consisted of single-gene screening (n = 21) and custom-designed hearing loss gene panel sequencing (n = 284) targeting known nonsyndromic and syndromic hearing loss genes in a diagnostic setup. Retrospective reanalysis of sequencing data was conducted by applying the current American College of Medical Genetics and Genomics/Association for Molecular Pathology guidelines.
Results:
A genetic diagnosis was established for 75 (25%) of the probands that involved 75 causal variants in 35 genes, including 16 novel causal variants and 9 medically significant variant reclassifications. Nearly half of the solved cases (47%; n = 35) were related to variants in the five most frequently affected genes: GJB2 (25%), MYO15A, WFS1, SLC26A4, and COL11A1 (all 5%). Nearly one-quarter of the cases (23%; n = 17) were associated with variants in seven additional genes (TMPRSS3, COL4A3, LOXHD1, EDNRB, MYO6, TECTA, and USH2A). The remaining one-third of single cases (33%; n = 25) were linked to variants in 25 distinct genes. Diagnostic rates and gene distribution were highly dependent on phenotypic characteristics. A positive family history of autosomal-recessive inheritance in combination with early onset and higher grades of hearing loss significantly increased the solve rate up to 60%, while late onset and lower grades of hearing loss yielded significantly fewer diagnoses. Regarding genetic diagnoses, autosomal-dominant genes accounted for 37%, autosomal-recessive genes for 60%, and X-linked genes for 3% of the solved cases. Syndromic/nonsyndromic hearing loss mimic genes were affected in 27% of the genetic diagnoses.
Conclusions:
The genetic epidemiology of the largest German cohort subjected to comprehensive targeted sequencing for hereditary hearing loss to date revealed broad causal gene and variant spectra in this population. Targeted hearing loss gene panel analysis proved to be an effective tool for ensuring an appropriate diagnostic yield in a routine clinical setting including the identification of novel variants and medically significant reclassifications. Solve rates were highly sensitive to phenotypic characteristics. The unique population-adapted and balanced age distribution of the cohort favoring late hearing loss onset uncovered a markedly large contribution of autosomal-dominant genes to the diagnoses which may be a representative for other age balanced cohorts in other populations.
Keywords: Deafness, Gene panel diagnostics, Genetic epidemiology of German hearing loss patients, Hearing loss genetic diagnostics, Hereditary hearing loss, Syndromic/nonsyndromic hearing loss mimic genes
Introduction
Hearing loss is the most prevalent sensory disorder, negatively affecting communication, social interaction, and quality of life, and worldwide is the third leading cause of disability (Cunningham & Tucci 2017; World Health Organization 2018; GBD Hearing Loss Collaborators 2021). A commonly cited figure of the World Health Organization (WHO) is that 6.1% of the world’s population (466 million people) are affected by disabling hearing loss, defined as hearing impairment above 40 dB HL (for adults ≥15 years of age) and 30 dB HL (for children from birth to 14 years of age) in the better ear (World Health Organization 2018; Olusanya et al. 2019b). However, the definition of disabling hearing loss excludes the clinically significant entities of all unilateral (<20 dB HL in the better ear; ≥35 dB HL in the worse ear) and mild (20–34.9 dB HL) bilateral hearing impairment (Global Burden of Disease Hearing Loss Expert Group et al. 2013; Olusanya et al. 2019a). Accounting for unilateral and mild bilateral hearing loss as suggested by the Global Burden of Disease (GBD) Hearing Loss Expert Group, the global hearing loss prevalence is much higher and was projected to be 18.7% of the world’s population (1.4 billion people) in 2015 (GBD 2015 Disease and Injury Incidence and Prevalence Collaborators 2016; Tucci et al. 2017; Wilson et al. 2017; James et al. 2018; Olusanya et al. 2019b). Four years later and according to the current figures of the GBD 2019 Hearing Loss Collaborators and the WHO World Report on Hearing, 20.3% (1.57 billion people) of the world’s population or “1 in 5” had mild to complete (≥20 dB HL) hearing loss in 2019 (GBD Hearing Loss Collaborators 2021; Wilson & Tucci 2021; World Health Organization 2021).
In Germany, the prevalence of hearing loss, including mild hearing loss, in the adult population has been estimated to be 15.5% according to WHO criteria (von Gablenz et al. 2020). This prevalence is slightly lower than that reported in other European and U.S. studies with similar dates of completion and applied parameters (von Gablenz et al. 2017). Recent meta-analyses have shown a hearing loss prevalence of 16% to 25% for adults (Löhler et al. 2019) and 1% to 4% for children and adolescents (Schmucker et al. 2019) in Germany.
Hereditary hearing loss accounts for the major portion of hearing loss throughout all age groups. It is classified according to clinical phenotype, including the pattern of inheritance, distinctive physical examination results, and audiologic features that may suggest nonsyndromic or syndromic types of hearing loss, as well as according to the genotype determined by causal variants in hearing loss-associated genes. Hereditary hearing loss exhibits a variable age of onset, grade, and progression depending on the underlying genes and variants involved. However, even in patients with identical genotypes, hereditary hearing loss may exhibit broad heterogeneity of the clinical phenotype. It is assumed that hereditary hearing loss follows an autosomal-recessive mode of inheritance in 80% of cases, with 19% following an autosomal-dominant and 1% an X-linked mode of inheritance (Shearer et al. 2017). In congenital hearing loss onset, a genetic background accounts for up to 80% of cases (Korver et al. 2017; Shearer et al. 2017). In childhood onset, genetic causes account for 60% of hearing loss cases in developed countries (Morton & Nance 2006). Hereditary hearing loss appearing in childhood, adolescence, and early and middle adulthood is mainly determined by monogenic factors with complete penetrance of causative genetic variants in a single gene. However, during adulthood, hearing loss increasingly develops from the multifactorial interaction of environmental and polygenic factors in more than one gene that shape a complex disease situation, especially for age-related hearing loss in late adulthood (Vuckovic et al. 2018). Nevertheless, the contribution of genetic factors even in age-related hearing loss persists at an estimated rate of 50%, and this also includes monogenic forms (Boucher et al. 2020; Van Eyken et al. 2007a,b).
Presently, 123 genes are known to be associated with nonsyndromic hearing loss (NSHL) (Azaiez et al. 2018; Van Camp & Smith 2021) and over 600 recognized hereditary syndromes include hearing loss as part of the clinical synopsis (Parker & Bitner-Glindzicz 2015). In many of these syndromes, hearing loss is the first clinical symptom, which has led to their designation of affected genes as NSHL mimics (Sloan-Heggen et al. 2016; Shearer et al. 2019). Widespread genetic testing for hearing loss was initiated more than two decades ago after the discovery of GJB2 as the first autosomal-recessive hearing loss-associated gene (Kelsell et al. 1997). In the 2000s, single-gene screening for GJB2 became the most frequent genetic test for hearing loss owing to the substantial genetic load of causal GJB2 variants in several populations worldwide and the relative ease of sequencing GJB2, a small, single-exon gene (Smith et al. 2005; Snoeckx et al. 2005). Technologically, for more than a decade, genetic testing remained constrained to targeted single-gene Sanger sequencing with a relatively low diagnostic yield. The discovery of more than 40 new hearing loss-associated genes over the course of the 2000s led to continuous growth in the genetic heterogeneity available for diagnostic evaluation. This growth led to a diagnostic predicament of how to choose the next single causative gene after GJB2 was ruled out (Hilgert et al. 2009).
Completion of the Human Genome Project in 2003 (Collins et al. 2003) and the advent of next-generation sequencing (NGS) (Tucker et al. 2009) represented great leaps forward in genetic testing. This technical progress eliminated the single-gene roadblock and enabled the development of targeted hearing loss gene panels, which were first described and introduced to the clinic in 2010 (Shearer et al. 2010). Targeted hearing loss panels including a substantial number of or possibly all known hearing loss genes quickly became established as the new standard of care (Shearer & Smith 2012; Alford et al. 2014; Shearer & Smith 2015). Genetic hearing loss panels are now offered by multiple laboratories in many countries, as outlined in the NIH genetic testing registry (Rubinstein et al. 2013). Since NGS has also significantly facilitated novel gene discovery, the number of hearing loss genes has continuously expanded, entailing the repeated addition of novel genes to updated versions of targeted hearing loss panels. Over the last decade, the number of validated genes available for clinical diagnostics has tripled, with more than 200 known genes currently used in routine clinical genetic testing in some centers (Rubinstein et al. 2013; Thorpe & Smith 2020).
Hearing loss gene panels have a proven cost-effectiveness and high diagnostic yield of up to 40% across all ages, hearing loss grades and ethnicities (Sloan-Heggen et al. 2016). Depending on the number and selection of genes included, phenotypic characteristics, the ethnic background of the patient population studied, and the variant interpretation rules applied, a wide range of diagnostic yields has been uncovered (Shearer et al. 2019; Vona et al. 2019). To date, for the German population, only a small-scale report including 30 probands describing the diagnostic yield of gene panel sequencing based on 80 or 129 genes in two different panel versions has been published (Vona et al. 2014). Based on this small cohort, reliable insight into the molecular genetic epidemiology of German patients is not feasible. Another multicenter study including 131 Western European probands with presumed autosomal-recessive NSHL from two centers in Belgium and Germany estimated that combined prescreening for GJB2 followed by targeted panel sequencing using a panel of 79 genes would result in a projected genetic diagnosis in a total of 25% to 30% of analyzed patients and a 10% increase in sensitivity over that of GJB2 testing alone (Sommen et al. 2016). Here, we report on a combined targeted sequencing approach applied to 305 patients recruited between 2011 and 2018 at a single center (Hearing Center; Department of Otolaryngology—Head & Neck Surgery, Tübingen, Germany) that resulted in genetic diagnosis in 25% of the patients according to the current American College of Medical Genetics and Genomics/Association for Molecular Pathology (ACMG/AMP) guideline criteria and hearing loss expert panel (HL-EP) specifications (Richards et al. 2015; Oza et al. 2018).
Materials and Methods
Patient Recruitment
This retrospective study was approved by the Ethics Committee at the University of Tübingen (Institutional Review Board; IRB). Written informed consent was obtained from patients or, in the case of minors, from their legal guardians. The patients included in this study had been sequentially enrolled in genetic diagnostic testing over the course of eight years (2011–2018) without phenotype restriction, including all ages. Only one proband is represented for each family. Relatives of patients were not included in this analysis, except for segregation, as stated. Otherwise, no exclusions were made. Phenotypic data, family history, and ethnicity were recorded. Ethnicity was assigned by geographic descent according to the United Nations “Standard Country or Area Codes for Statistical Use” originally published as Series M, No. 49 (M49 standard) (Statistics Division United Nations 2021). The involved area codes were Western European (M49 code 155), Eastern European (M49 code 151), Southern European (M49 code 039), Western Asian (M49 code 145), and Southern Asian (M49 code 034).
Statistical Analysis
Clinical and audiologic phenotypic data were recorded for all probands of the cohort. Diagnostic rates for specific phenotypic features (inheritance, hearing loss age of onset, hearing loss grade, laterality, and ethnicity) were compared for a specific group to all other members of the cohort using Fisher’s exact test (levels of significance: p < 0.05; p < 0.005). Data were compiled using Microsoft Excel and analyzed using IBM SPSS Statistics V24.
Testing Workflow
Genomic DNA extracted from EDTA-treated blood (3–5 mL) was isolated according to the manufacturers’ recommendations by using either a QIAamp DNA Blood Maxi Kit or a QIAsymphony instrument (Qiagen, Hilden, Germany) for all sequencing. DNA quantity and quality were determined using both a Qubit fluorometer and a NanoDrop ND-8000 (Thermo Fisher Scientific, Dreieich, Germany). Genetic testing was applied to a total of 305 hearing-impaired individuals/families with a suspected genetic hearing loss etiology. Inheritance patterns were based on family history. The genetic testing workflow consisted of single-gene screening (n = 21) and custom-designed hearing loss gene panel sequencing (n = 284) targeting known nonsyndromic and syndromic hearing loss genes for a total of 305 probands.
Single-gene Screening
Depending on the clinical phenotype, selected single-gene analysis by Sanger sequencing was performed for the genes GJB2 (n = 18), GJB3 (n = 3), GJB6 (n = 3), OTOF (n = 1), and SLC26A4 (n = 2) in 21 patients. Single-gene analysis was used as a screening tool at the beginning of the study (2011–2016), followed by targeted gene panel analysis in unsolved cases. After July 2016, the single-gene sequencing step was omitted, and genetic analysis directly proceeded to comprehensive gene panel sequencing including all genes previously employed in the single-gene screening.
Targeted Gene Panel Design, Sequencing, and Bioinformatic Analysis
Targeted sequencing of genes known to be associated with nonsyndromic and syndromic hearing loss was performed in 284 patients with five panel versions over a period of eight years (see Table in Supplemental Digital Content 1, http://links.lww.com/EANDH/A958, which shows the gene content included in each panel version, and Table in Supplemental Digital Content 2, http://links.lww.com/EANDH/A959, which shows all gene transcripts used for gene panel design). These hearing loss panels were sequentially numbered version 1 (n = 2 tested patients), version 2 (n = 100), version 3 (n = 44), version 4 (n = 127), and version 5 (n = 11). The versions of the hearing loss panel evolved in cooperation with the Center for Genomics and Transcriptomics (CeGaT) GmbH, Tübingen, Germany, and were expanded over an 8-year period from 2011 to 2018, with an increasing total number of hearing loss genes in each version. Version 1 (October 2011–April 2012) covered 71 genes, version 2 (May 2012–May 2013) 95 genes, version 3 (June 2013–November 2014) 110 genes, version 4 (December 2014–September 2017) 164 genes, and Version 5 (October 2017–2018) 179 genes.
Library preparation targeting the coding and flanking intronic sequences was performed using an Agilent in-solution target enrichment procedure. Libraries were sequenced using the SOLiD 5500xl System (n = 95; Applied Biosystems, Thermo Fisher Scientific, Waltham, MA/Life Technologies, Grand Island, NY) or the Illumina HiSeq 2500 (n = 155), HiSeq 4000 (n = 16), or NovaSeq 6000 (n = 18) system (Illumina, San Diego, CA). The enrichment kit and method used were established, validated, and provided by CeGaT GmbH. CeGaT is accredited by the German Accreditation Body (DAkkS) according to DIN EN ISO 15189:2014, by the College of American Pathologists and is CLIA certified as previously described (Weisschuh et al. 2020).
The 2 × 75 base pair (bp) reads produced by the SOLiD 5500xl platform were analyzed using LifeScope software version 2.1 or 2.5 (Life Technologies). Reads were mapped to the human reference genome (hg19) with a BLAST-like mapping algorithm using color codes. Single-nucleotide variants and small insertions/deletions (indels) were called with a frequentist algorithm at high-coverage positions or with a Bayesian algorithm. Sequencing reads generated by the Illumina platforms were demultiplexed using Illumina CASAVA (1.8.2) (Illumina). Adapter sequences were removed with Skewer 0.1.116, and the trimmed reads were mapped to the human reference genome (hg19) using Burrows–Wheeler Aligner (BWA-mem 0.7.2) (Li & Durbin 2010). Reads mapping to more than one location with identical mapping scores were discarded (internal software). Read duplicates resulting from PCR amplification were removed (SAMtools 0.1.18). Variants were called using SAMtools and VarScan (2.3.3) (http://dkoboldt.github.io/varscan). Technical artifacts were removed (custom-made software), and the remaining variants were annotated based on several internal and external databases. Since November 2012 (during version 2), copy number variations (CNVs) have been computed using an in-house-developed method (by CeGaT) based on sequence coverage depth as previously described (Dohrn et al. 2017). All analyzed panel genes were part of the CNV analysis, with the exception of STRC in panel version 3 (see Table in Supplemental Digital Content 1, http://links.lww.com/EANDH/A958). Starting with gene panel version 3 (June 2013), screening included known deep intronic variants in genes such as USH2A, CDH23 and CLRN1. Version 3 (June 2013–November 2014) and version 4 (December 2014–September 2017) contained: USH2A, rs1421396656; CDH23, rs1183500485; version 5 (October 2017–2018): USH2A, 8 intronic positions; CDH23, rs1183500485 and rs367928692. The current version 6 (since January 2021) includes deep intronic variants for USH2A: 26 intronic positions, CDH23: rs1183500485, rs367928692 and CLRN1: rs924764326, rs373838930, and rs36798692.
Variant Filtering and Classification
Only variants (single-nucleotide variants/small indels) in the coding and flanking intronic regions (8 bp) with a minor allele frequency (MAF) <1.5% were evaluated using prediction tools to predict the effects of single-nucleotide variants or small indels on splicing within this splice region. Known pathogenic deep intronic variants and known disease-causing variants (according to a licensed version of the Human Gene Mutation Database (HGMD), HGMD Professional version 2020.4) located within ±30 bp of flanking intronic regions and having MAFs of up to 5% were also evaluated. MAFs were taken from the following databases: 1000 Genomes, dbSNP, Exome Variant Server, ExAC, gnomAD, and an in-house database. At least one causative or rare variant was resequenced using conventional Sanger sequencing, providing a second, independent confirmation of the variant and confirming that the NGS data and stock DNA matched. Depending on the quality of the NGS data, causative variants were validated using Sanger sequencing. Individual medical reports for all patients contained all variants with an MAF <1% (in genes associated with an autosomal-recessive mode of inheritance) or <0.1% (in genes associated with an autosomal-dominant mode of inheritance) and excluded variants classified as likely benign (LB) or benign (B) according to the current literature at the time of sequencing.
A retrospective reanalysis of all sequencing data was conducted uniformly, applying current knowledge and criteria. In silico prediction of variants was performed based on the output of the programs MutationTaster2 (Adzhubei et al. 2010), PolyPhen-2, SIFT (Kumar et al. 2009) assisted by Alamut (Interactive Biosoftware; SOPHiA GENETICS, Rouen, France). To assess the effects of variants on splicing, multiple tools were used (Jian et al. 2014) and were complemented with additional in silico predictions in NetGene2 (Brunak et al. 1991; Hebsgaard et al. 1996) and Fruitfly (Celniker et al. 2002).
Public archives of interpretations, namely, an archive of clinically relevant variants (ClinVar) (Harrison et al. 2016; Landrum et al. 2016, 2020), the Genome Aggregation Database (gnomAD) (Karczewski et al. 2020), the Deafness Variation Database (DVD) (Shearer et al. 2014a; Azaiez et al. 2018) and a licensed version of the HGMD (HGMD Professional, version 2020.4) (Stenson et al. 2020), were used to assist in variant prioritization and reanalysis of all data sets based on current versions.
Based on this reanalysis, variants were classified according to ACMG/AMP guidelines for genetic hearing loss (Richards et al. 2015; Oza et al. 2018) assisted by the public version of The Human Genomic Variant Search Engine (VarSome) (Kopanos et al. 2019). According to these recommendations, variants reported in this study were categorized as follows: pathogenic (P), likely pathogenic (LP), variants of uncertain significance (VUSs), LB, and B.
While the original diagnostic reports written by “Praxis für Humangenetik” addressed to the referring physician used a different grading system, the following criteria were applied to reach a genetic diagnosis to qualify as a “solved” case as described previously (Weisschuh et al. 2020): (1) one heterozygous variant classified as “P” or “LP” was detected in a gene categorized for autosomal-dominant hearing loss; (2) a hemizygous variant classified as “P” or “LP” was found in a gene categorized for X-linked hearing loss; (3) two variants (suspected or shown to lie on separate alleles) classified as “P” or “LP” were identified in a gene categorized to cause autosomal-recessive hearing loss. Detection of a single “P” or “LP” allele in a gene associated with autosomal recessive disease either alone or in combination with a “VUS” variant was classified as an “unsolved” case.
Results
Patient Cohort Summary and Audiologic Characterization
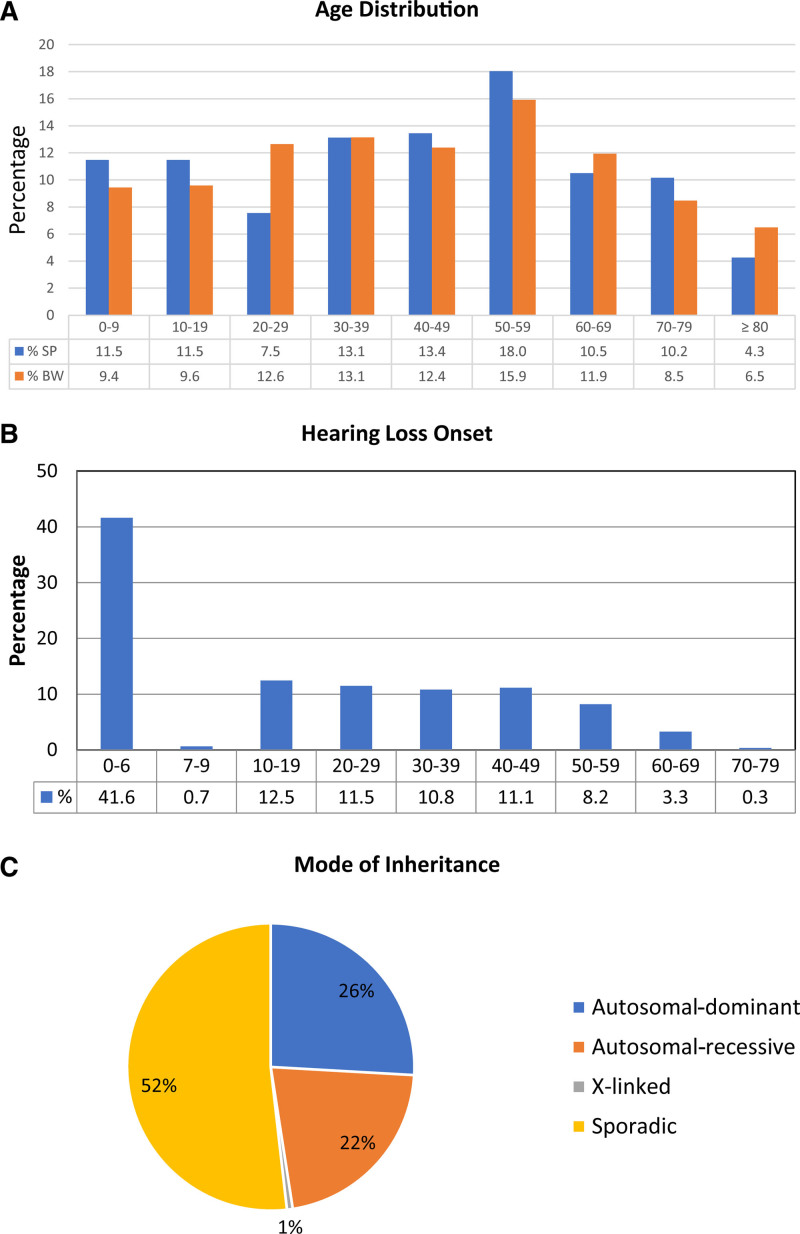
The patient recruitment area of the Hearing Center Tübingen is geographically located in the center of Southwest Germany (State of Baden-Württemberg; BW). In the study, 305 unrelated hearing-impaired probands well-balanced across all age groups (mean age; x̅ ± SD: 42.4 ± 23.6; year 2018) were included (Fig. 1A; Table 1; see Table in Supplemental Digital Content 3, http://links.lww.com/EANDH/A960). The mean age of the cohort was close to the mean age of the population in Southwest Germany on the same reference date (mean age: 43.5; year 2018) (State Statistical Office Baden-Württemberg 2021). The age structure of the cohort shows that probands were evenly distributed across all age groups (≈11%/decade), with a peak in the sixth decade and a decline thereafter. Similar to mean age, the age structure of the cohort is highly representative of the age distribution in the general population of Southwest Germany (State Statistical Office Baden-Württemberg 2021) (Fig. 1A). The cohort comprised 149 males (48.9%; state of BW: 49.7%, year 2018) and 156 females (51.1%; state of BW: 50.3%, year 2018), also a representative of the gender distribution in the population of Southwest Germany (State Statistical Office Baden-Württemberg 2021) (Table 1; see Table in Supplemental Digital Content 3, http://links.lww.com/EANDH/A960).
Fig. 1.
Demographic and phenotypic characteristics of the cohort. A, Age structure of the study population (SP) (n = 305) across all age groups in decades (blue bars). The percentage for each decade is shown below the bar. The age structure of the cohort was compared to the age distribution in Southwest Germany (BW) for the same reference year (2018) and decades (data from State Statistical Office Baden-Württemberg 2021) (orange bars). B, Age-related distribution of hearing loss onset. The phases of prelingual and perilingual hearing loss onset, including congenital onset, are shown in the first bar (≤6 years). Postlingual hearing loss onset starts at the second bar (7–9 years) and continues for subsequent decades. The percentage for each time window is shown below the bar. C, Overview of the modes of inheritance based on family history in the total patient cohort. Pie chart showing the respective proportions of autosomal-dominant, autosomal-recessive, and X-linked inheritance patterns, as well as patients with sporadic hearing impairment.
TABLE 1.
Demographic characteristics and ethnicity of the patient cohort
| Characteristic | Number | % |
|---|---|---|
| Age (mean ± SD), yrs | 42.4 ± 23.6 | |
| Sex | ||
| Male | 149 | 49 |
| Female | 156 | 51 |
| All | 305 | 100 |
| Ethnicity | ||
| Western European | 237 | 77.8 |
| Western Asian | 31 | 10.2 |
| Eastern European | 17 | 5.6 |
| Southern European | 14 | 4.6 |
| Southern Asian | 5 | 1.6 |
| Other | 1 | 0.3 |
Similar to the balanced age distribution, phases of hearing loss onset from congenital onset to late adult onset in the seventh decade were disseminated over all age groups in the cohort. The phases of congenital, prelingual and perilingual hearing loss onset are defined until 6 years of age (age ≤ 6 years; early onset), representing a critical developmental time period that is essential for auditory plasticity (Sharma et al. 2002) and the acquisition of language and music perception capabilities (Fuller et al. 2013, 2019). Postlingual hearing loss onset is defined as starting after this time period (age > 6 years; late onset). Pre/perilingual hearing loss, including congenital hearing loss onset (≤6 years; early onset), was recognized in 42% (n = 127) and postlingual onset (>6 years; late onset) in 58% (n = 178) of the probands. In the postlingual group, the prevalence of hearing loss onset was almost equal among the second to fifth decades (≈11.5%/decade) and started to decline in the sixth decade (Fig. 1B).
Based on family history, the pattern of inheritance included 66 patients (22%) with autosomal-recessive hearing loss, 79 patients (26%) with autosomal-dominant hearing loss, two male patients (1%) with X-linked hearing loss and 158 patients (52%) with a sporadic type of hearing loss (Table 2; Fig. 1C; see Table in Supplemental Digital Content 3, http://links.lww.com/EANDH/A960).
TABLE 2.
Family history in relation to hearing loss onset and progression
| Family History | Total | Early Onset | Late Onset |
|---|---|---|---|
| Autosomal-recessive | 66 (22%) | 38 (57%) | 28 (43%) |
| Stable | 26 (68%) | 6 (21%) | |
| Progressive | 12 (32%) | 22 (79%) | |
| Autosomal-dominant | 79 (26%) | 21 (27%) | 58 (73%) |
| Stable | 8 (38%) | 4 (7%) | |
| Progressive | 13 (62%) | 54 (93%) | |
| X-linked | 2 (1%) | 1 (50%) | 1 (50%) |
| Stable | 0 (0%) | 0 (0%) | |
| Progressive | 1 (100%) | 1 (100%) | |
| Sporadic | 158 (52%) | 66 (42%) | 92 (58%) |
| Stable | 39 (59%) | 19 (21%) | |
| Progressive | 27 (41%) | 73 (79%) |
Early hearing loss onset refers to congenital/pre/perilingual onset (≤6 years of age).
Late hearing loss onset refers to postlingual onset (>6 years of age).
For the autosomal-recessive inheritance group, an early hearing loss onset appeared in the majority of cases (n = 38; 57%). Hearing loss was stable in 26 (68%) and progressive in 12 (32%) of these early onset cases. Late hearing loss onset was observed in 28 (42%) of the autosomal-recessive cases. Hearing loss was stable in 6 (21%) and progressive in 22 (79%) of these postlingual cases (Table 2).
In the autosomal-dominant inheritance group, hearing loss onset was recognized in the early onset time period in one-fourth of the cases (n = 21; 27%). Hearing loss was stable in 8 (38%) and progressive in 13 (62%) of these early onset cases. Late hearing loss onset was observed in 58 (73%) of the autosomal-dominant cases. Hearing loss was stable in 4 (7%) and progressive in 54 (93%) of these postlingual cases (Table 2).
The distribution in the sporadic group presented 66 (42%) early and 92 (58%) late hearing loss onset cases. In the early onset group, 39 (59%) patients showed stable hearing loss, and 27 (41%) showed progressive hearing loss. In the late onset group, 19 (21%) showed stable and 73 (79%) showed progressive hearing loss (Table 2).
Of the two cases of male X-linked hearing loss, one displayed progressive early hearing loss onset, and the other progressive postlingual hearing loss onset.
Grades of hearing impairment were classified as recommended by the GBD Expert Group on Hearing Loss (Global Burden of Disease Hearing Loss Expert Group et al. 2013; Olusanya et al. 2019a; GBD Hearing Loss Collaborators 2021). The results for all single ears (n = 610) were distributed among all hearing loss grades, including normal hearing (−10.0 to 19.9 dB HL) (n = 48; 7.9%), mild hearing loss (20.0–34.9 dB HL) (n = 43; 7.0%), moderate hearing loss (35.0–9.9 dB HL) (n = 69; 11.3%), moderately severe hearing loss (50.0–64.9 dB HL) (n = 81; 13.3%), severe hearing loss (65.0–79.9 dB HL) (n = 82; 13.4%), profound hearing loss (PHL; 80.0–94.9 dB HL) (n = 78; 12.8%), and complete or total hearing loss (CHL; ≥95.0 dB HL) (n = 209; 34.3%) (Fig. 2A).
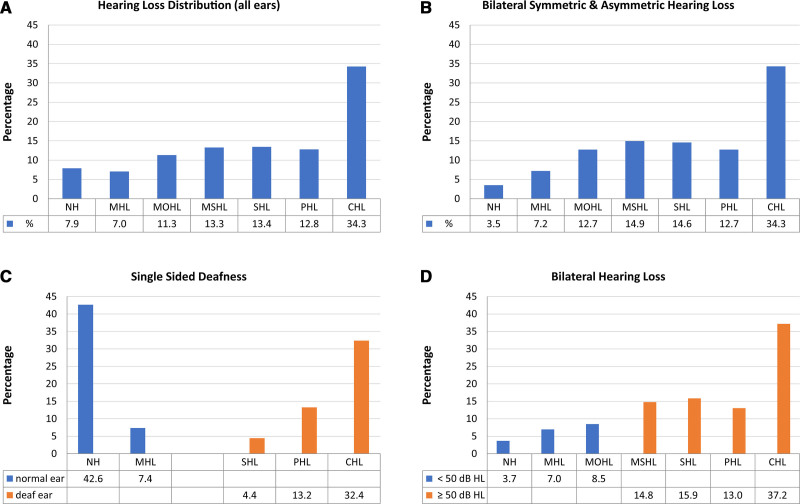
Fig. 2.
Distribution of hearing loss grades in the cohort. A, Distribution of hearing loss grades for all single ears (n = 610). B, Distribution of hearing loss grades for probands with bilateral symmetric (n = 241) and asymmetric (n = 30) hearing loss (AHL) (better ear: PTA >30 and ≤55 dB HL; poorer ear PTA ≥70 dB HL; interaural threshold gap ≥15 dB HL). C, Distribution of hearing loss grades for probands with single-sided deafness (SSD) (n = 34); better ear (blue bars): PTA ≤30 dB HL; poorer ear (orange bars): PTA ≥70 dB HL; interaural threshold gap: ≥40 dB HL; according to the consensus framework for SSD and AHL (Van de Heyning et al. 2016). D, Separation of bilateral mild and moderate (<50 dB HL) hearing loss (blue bars) (n = 88) and higher-grade (≥50 dB HL) bilateral hearing loss (orange bars) (n = 372) for single ears. NH, normal hearing (−10.0 to 19.9 dB HL); MHL, mild hearing loss (20.0 to 34.9 dB HL); MOHL, moderate hearing loss (35.0 to 49.9 dB HL); MSHL, moderately severe hearing loss (50.0 to 64.9 dB HL); PHL, profound hearing loss (80.0 to 94.9 dB HL); SHL, severe hearing loss (65.0 to 79.9 dB HL); CHL, complete or total hearing loss (≥95.0 dB HL). Classification follows the recommendations of the Global Burden of Disease (GBD) Expert Group on Hearing Loss (Global Burden of Disease Hearing Loss Expert Group et al. 2013; Olusanya et al. 2019a). Percentage values for different hearing loss grades are shown below bars.
With regard to laterality, the majority of probands presented with bilateral symmetric hearing loss (n = 241; 79.0%). One-tenth displayed bilateral asymmetric hearing loss (AHL) (n = 30; 9.8%) (better ear: PTA >30 and ≤55 dB HL; poorer ear PTA ≥70 dB HL; interaural threshold gap ≥15 dB HL) as defined by the consensus framework for single-sided deafness (SSD) and AHL (Van de Heyning et al. 2016) (Fig. 2B).
A further tenth exhibited unilateral hearing loss within the definition of SSD (n = 34; 11.1%) (better ear: PTA ≤ 30 dB HL; poorer ear: PTA ≥ 70 dB HL; interaural threshold gap: ≥ 40 dB HL) (Van de Heyning et al. 2016) (Fig. 2C).
Lower grades of bilateral hearing loss, including normal, mild, and moderate (<50 dB HL) hearing loss (n = 88; 19.1%), were much less frequent than higher grades (≥50 dB HL) of bilateral hearing loss (n = 372; 80.9%) (Fig. 2D).
The ethnic background of the probands was primarily Western European (n = 237; 77.8%), with the majority coming from the local region of Southwest Germany. Other ethnicities (n = 68; 22.2%) representing the general migration background of the regional population were Western Asian (n = 31; 10.2%), Eastern European (n = 17; 5.6%), Southern European (n = 14; 4.6%), Southern Asian (n = 5; 1.6%), and other (n = 1; 0.3%) (Table 1; Fig. 3; see Table in Supplemental Digital Content 3, http://links.lww.com/EANDH/A960). This distribution is based on a slightly higher general migration background in the population in Southwest Germany (33.4%; year 2019; 2018 data were not available for this comparison) (State Statistical Office Baden-Württemberg 2021).
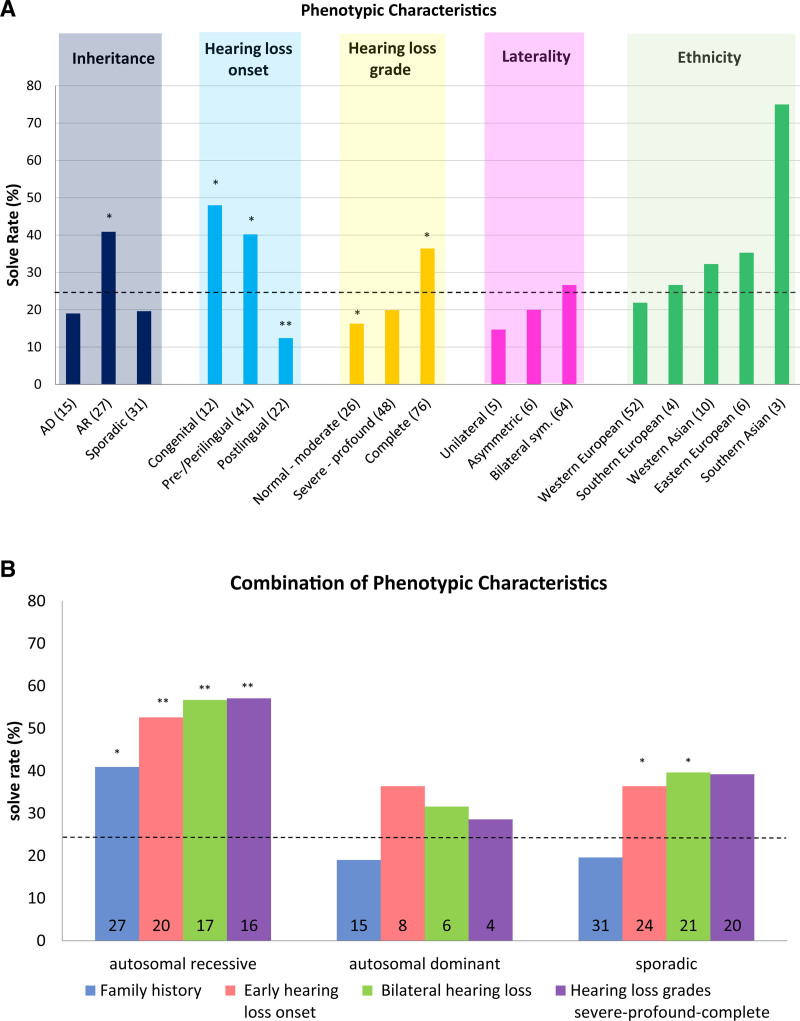
Fig. 3.
Impacts of phenotypic characteristics on diagnostic solution rates. A, Individual clinical and audiologic characteristics impact the diagnostic rates. Color shading indicates five clinical and audiologic categories: inheritance, hearing loss onset, hearing loss grade, laterality, and ethnicity. The dashed line indicates the overall diagnostic rate for this study (25%). The bars in the column chart indicate the percentages of patients with the noted individual characteristics in the five categories (the number of patients is noted for each characteristic). Statistical significance was determined with Fisher’s exact test (*p < 0.05 and **p < 0.005). B, Combination of above-average clinical and audiological characteristics impacts the diagnostic rates. Sequential addition of phenotypic criteria to inheritance leads to a stepwise increase in the diagnostic rate (the number of patients is noted for each characteristic). Statistical significance was determined with Fisher’s exact test (*p < 0.05; **p < 0.005). Adapted from Sloan-Heggen et al. (2016).
Diagnostic Yield of Targeted Genetic Sequencing and Phenotypes
The overall diagnostic yield for the cohort of 305 unrelated patients who underwent combined targeted genetic testing was 25% (n = 75) according to ACMG/AMP criteria (Richards et al. 2015) and the hearing loss specification according to the HL-EP (Oza et al. 2018). Segregation testing was performed in 18 of the solved cases (see Table in Supplemental Digital Content 4, http://links.lww.com/EANDH/A961).
The diagnostic rate varied with phenotypic categories such as inheritance, hearing loss onset, hearing loss grade, laterality, and ethnicity (Fig. 3A).
In probands with a family history of autosomal-recessive hearing loss, the solve rate was significantly higher than average (41%; n = 27/66; p < 0.05). In patients with a family history of autosomal-dominant [19%; n = 15/79; not significant (n.s.)] or sporadic (20%; n = 31/158; n.s.) hearing loss, the solve rate was lower but not significantly so.
For hearing loss onset, the diagnostic rate increased significantly for congenital (48%; n = 12/25; p < 0.05) and pre/perilingual (40%; n = 41/102; p < 0.05) hearing loss and declined significantly for postlingual hearing loss onset (12%; n = 22/178; p < 0.005). Combined congenital and pre/perilingual hearing loss onset (early onset) also significantly increased the diagnostic rate (42%; n = 53/127; p < 0.05).
Considering the hearing loss grade for single ears, cases with complete hearing loss yielded a significantly higher solve rate (36%; n = 76/209; p < 0.05). Lesser hearing loss grades in the severe-to-profound range (20%; n = 48/241; n.s.) and in the normal to moderate range (16%; n = 26/160; p < 0.05) showed a continuous trend toward lower solve rates.
Hearing loss laterality did not significantly affect the solve rate; however, the trend indicated that patients with symmetric bilateral hearing loss (27%; n = 64/241; n.s.) were more likely to receive a genetic diagnosis than patients with asymmetric bilateral hearing loss (20%; n = 6/30; n.s.) or SSD (unilateral hearing loss) (15%; n = 5/34; n.s.).
Ethnicity did not significantly influence the diagnostic rate. As stated above, the cohort was dominated by Western European probands (77.8%; n = 237/305). Their diagnostic rate was slightly below average (22%; n = 52/237; n.s.). Other ethnicities showed considerable variation in solve rates, but the number of diagnosed probands in these ethnic groups was too small to reach significance.
The combination of clinical and audiological phenotypic characteristics starkly impacted the solve rates. Inheritance patterns for autosomal-recessive, autosomal-dominant and sporadic hearing loss were combined with other clinical characteristics. The stepwise combination of the three inheritance patterns with other characteristics such as early hearing loss onset (congenital and pre/perilingual), bilateral symmetric laterality and high hearing loss grades (severe, profound, and complete) led to an increase in solve rates to over 60% for all inheritance patterns. For the autosomal-recessive and sporadic inheritance patterns, the increase in diagnostic rates was significant (p < 0.005) (Fig. 3B).
Gene Distribution
The diagnosis of the 75 solved cases involved 35 genes and 70 clinically relevant (P, LP) variants (Fig. 4; see Table in Supplemental Digital Content 4, http://links.lww.com/EANDH/A961, for detailed gene and variant distributions of solved cases).
Fig. 4.
Distribution of 35 genes among 70 clinically relevant (pathogenic, likely pathogenic) variants among 75 solved cases. A, Pie chart of the overall gene distribution. B, Pie chart of the gene distribution for autosomal-recessive genes. C, Autosomal-dominant genes. D, Variant distribution for all cases (n = 305) of 443 different variants from a total of 106 different genes. Classification as pathogenic, likely pathogenic, variants of unknown significance, likely benign and benign according to ACMG/AMP criteria for hearing loss (Oza et al. 2018; Richards et al. 2015).
The overall gene distribution showed that nearly half of the solved cases (47%; n = 35) were related to variants in the five most frequently affected genes: GJB2 (25%), MYO15A, WFS1, SLC26A4, and COL11A1 (each 5%). Nearly one-quarter of the cases (23%; n = 17) were correlated to causative variants in seven additional genes, namely, TMPRSS3, COL4A3, and LOXHD1 (each 4%; n = 3), and EDNRB, MYO6, TECTA, and USH2A (all 2.7%; n = 2). Overall, 69% of the solved cases (n = 52/75) were attributed to causative variants in only 12 genes. The remaining third (31%; n = 23) of the single cases were allocated to causative variants in 23 additional single genes (ACTG1, CDH23, COCH, COL2A1, COL4A5, COL9A2, DIAPH1, DIAPH3, EYA4, GATA3, KCNQ4, MANBA, MARVELD2, MITF, MYO7A, OTOF, PAX3, POU4F3, SMPX, SOX10, STRC, TFAP2A, and TMIE) (Fig. 4A; Supplemental Digital Content 4, http://links.lww.com/EANDH/A961, for gene distributions of solved cases).
After establishing a genetic diagnosis in the solved cases (n = 75/305; 25%), a group of probands received a genetic diagnosis that implicated a syndromic/NSHL mimic gene (20/75; 27%) (see Table in Supplemental Digital Content 5, http://links.lww.com/EANDH/A962 showing distribution of cases in syndromic/NSHL mimic genes). These included Waardenburg syndrome (n = 5; 6.3%), Alport syndrome (n = 4; 5.3%), Pendred syndrome (n = 4; 5.3%), Stickler syndrome (n = 3; 4%), Usher syndrome (n = 2; 2.7%), Branchio-Oculo-Facial syndrome (n = 1; 1.3%), and Beta-Mannosidosis (n = 1; 1.3%).
Gene Distribution According to Gene Associated Inheritance Pattern
After establishing a genetic diagnosis, the contribution of distinct genes was linked to their established mode of inheritance (i.e., autosomal-recessive genes, autosomal-dominant genes, and X-linked genes) (Fig. 4B and C). The list of candidate genes curated or precurated by the ClinGen Hearing Loss Gene Curation Expert Panel (HL GCEP) associated with nonsyndromic and syndromic hearing loss was used for classification (DiStefano et al. 2019). Genes with a dual mode of inheritance (COL4A3, MYO7A, TECTA, and WFS1) were classified on an individual case basis (see Table in Supplemental Digital Content 6, http://links.lww.com/EANDH/A963 showing HL GCEP classifications for all 35 genes that contributed to the diagnostic yield).
Autosomal-dominant genes accounted for 37% of the genetic diagnoses, autosomal-recessive genes for 60%, and X-linked genes for 3%.
At the single-gene level, causative variants in GJB2 were the most common cause among autosomal-recessive genes (45%), followed by MYO15A and SLC26A4 (both 10%), TMPRSS3 and LOXHD1 (both 7%). Among the autosomal-dominant genes WFS1 and COL11A1 (both 13%) appeared as the most frequent followed by EDNRB and MYO6 (both 6%) (see Fig. 4B and C, showing the distribution of autosomal-recessive and autosomal-dominant genes).
Gene Distribution in Early and Late Hearing Loss Onset
Hearing loss onset in the diagnosed cases (n = 75) was distributed between early onset (congenital and pre/perilingual stages) (72%; n = 54/75) (Fig. 5A), and the late onset (postlingual stages) (28%; n = 21/75) (Fig. 5B). In the larger early hearing loss onset group, 27 genes contributed to the diagnosis, while 16 genes participated in a diagnosis in the late onset group (Fig. 5A and B).
Fig. 5.
Gene distribution according to onset. Gene distribution of solved cases with (A) early (congenital and pre/perilingual hearing loss onset; ≤6 years) (27 genes) and (B) late (postlingual hearing loss onset; >6 years) (16 genes) hearing loss onset.
Early hearing loss onset primarily exhibited an autosomal-recessive mode of inheritance in more than a third of the cases (39%; n = 21/54). The other two-thirds were distributed among autosomal-dominant (15%; n = 8/54), sporadic (44%; n = 24/54), and X-linked (2%; n = 1/54) modes of inheritance. Late hearing loss onset was more evenly distributed among the autosomal-recessive (36%; n = 8/22), autosomal-dominant (27%; n = 6/22), and sporadic (32%; n = 7/22) modes of inheritance and one case of X-linked inheritance (4%; n = 1/22).
The distribution of the 27 genes associated with early onset was dominated by GJB2 (31%; n = 17), MYO15A (7%; n = 4), COL11A1, SLC26A4 (both: 6%; n = 3), associated with half of the cases (50%; n = 27). One-sixth of the cases (15%; n = 8) were associated with four genes, namely, WFS1, EDNRB, LOXHD1, and USH2A (all four: 4%; n = 2). The remaining one-third of single cases (35%; n = 19) were associated with 19 distinct genes (CDH23, COCH, COL2A1, COL9A2, DIAPH3, EYA4, MANBA, MARVELD2, MITF, MYO6, OTOF, PAX3, SMPX, SOX10, STRC, TECTA, TFAP2A, TMIE, and TMPRSS3) (Fig. 5A). Although these early onset cases were dominated by autosomal-recessive genes, nine autosomal-dominant genes also showed an association with early onset (COL11A1, EDNRB, COL2A1, DIAPH3, EYA4, MITF, PAX3, SOX10, and TFAP2A).
In the late onset group, the distribution of 16 genes showed predominance for COL4A3 (14%; n = 3), TMPRSS3, GJB2, and WFS1 (all: 9.5%; n = 2). Over half of the remaining diagnoses were represented by single cases (57%; n = 12) associated with 12 distinct genes (ACTG1, COL11A1, COL4A5, DIAPH1, GATA3, KCNQ4, LOXHD1, MYO6, MYO7A, POU4F3, SLC26A4, and TECTA) (Fig. 5B). Autosomal-dominant genes dominated this late hearing loss onset group. Four autosomal-recessive genes demonstrated an association with late hearing loss onset in single cases (TMPRSS3, GJB2, LOXHD1, and SLC26A4).
Variant Distribution, Classification, and Interpretation
In total, for all cases (n = 305), 443 different variants from 105 different genes were identified, demonstrating that the overall variant classification and interpretation was based on a much broader range of genes and variants than the genes (n = 35) and variants (n = 70) involved in the diagnostic yield of the solved cases (n = 75 of 305). Of these 443 variants, 25 (6%) were classified as P, 93 (21%) as LP, 304 (69%) as VUS, 15 (3%) as LB, and 6 (1%) as B according to ACMG/AMP criteria for hearing loss (Richards et al. 2015; Oza et al. 2018) (Fig. 4D).
In the solved cases, the clinically significant variants (P and LP) (n = 70) were differentially distributed across variant classifications, located in the coding regions, and canonical splice sites. Missense variants represented the majority of the variants (n = 28; 40%), followed by frameshift (n = 12; 17%), nonsense (n = 9; 13%), stop-gain (n = 6; 9%), splice donor (n = 7; 10%), splice acceptor (n = 3; 4%), missense and splice region (n = 2; 3%), and frameshift and splice region, deletion, and synonymous (all n = 1; 1%) variants (see Table in Supplemental Digital Content 4, http://links.lww.com/EANDH/A961 showing clinically significant variants types of all solved cases).
In addition, further clinically significant single heterozygous variants (P and LP) (n = 51) were found in unsolved cases (see Table in Supplemental Digital Content 7, http://links.lww.com/EANDH/A964 showing a list of heterozygous clinically significant variants types of undiagnosed patients). These heterozygous variants were located in various autosomal-recessive genes (n = 26) lacking a second causal variant. In these carrier patients, a genetic diagnosis could not be conclusively made since only one heterozygous P or LP variant was identified in genes associated with autosomal-recessive inheritance.
In the solved cases (n = 75), the ACMG/AMP variant classification deviated from the DVD and/or ClinVar classification for 45 (59%) of the identified clinically significant variants (P and LP) (n = 70). There was classification agreement for 31 (41%) variants. A comparative overview of the ACMG/AMP classification versus the ClinVar and DVD classifications for clinically relevant variants (P and LP) in the solved cases (n = 75) is given in Supplemental Digital Content 4, http://links.lww.com/EANDH/A961 (for variants in ClinVar with multiple submissions for pathogenicity, their most pathogenic submission was selected).
Novel Variants and Medically Significant Reclassifications
A substantial portion of the 46 deviating classifications was represented by 16 (35%) novel causal variants (within n = 13 families) previously listed in neither ClinVar nor DVD. These novel variants were distributed among the SLC26A4, TECTA, OTOF (all n = 2), SMPX, TMPRSS3, COL11A1, MYO6, MYO7A, EYA4, POU4F3, TFAP2A, KCNQ4, and GATA3 genes (all n = 1). Major categorical changes between ACMG/AMP and ClinVar/DVD that resulted in medically significant reclassifications were observed for 9 (20%) variants (within n = 9 families) of the 46 deviating classifications. These medically significant reclassifications were changes from VUS to LP in eight cases and from VUS to P in one case (the most pathogenic submission in either ClinVar or DVD was used for comparison). The 9 medically significant reclassifications were distributed among 8 genes (LOXHD1, n = 2; and COL4A3, COL9A2, COL11A1, EDNRB, MYO15A, WFS1, and DIAPH3, all n = 1). The remaining portion (n = 21; 46%) of the 46 variant classification discrepancies were medically nonsignificant reclassifications (i.e., P to LP). The pedigrees of all families and corresponding audiograms of all index patients with novel causal variants (n = 13 families) and medically significant reclassifications (n = 9 families) are shown in Supplemental Digital Content 8, http://links.lww.com/EANDH/A965.
In the group of novel causal variants, five cases were from simplex families, and eight cases were from multiplex families. In the group of medically significant reclassifications, six cases were from simplex families, and three cases were from multiplex families. In the multiplex families with novel causal variants, on average, 2.9 (range 2–6) family members were affected by hearing loss. In the multiplex families with medically significant reclassifications, 3.0 family members were affected by hearing loss.
Single-gene Sequencing
Until 2016, as a first screening step, targeted single-gene Sanger sequencing was applied for GJB2, GJB3, GJB6, SLC26A4, and OTOF, depending on the clinical and, if available, radiological presentation of the probands. Variant identification by single-gene sequencing solved 18 of 21 cases. The most frequently implicated gene was GJB2 (GJB2, n = 16; OTOF, n = 1; SLC26A4, n = 1) (see Table in Supplemental Digital Content 4, http://links.lww.com/EANDH/A961 for listing of variants). The variants detected in GJB2 (NM_004004.5) in a homozygous state (11/16) were c.35delG (7/16), c.71G>A (2/16), c.−23+1G>A (1/16), and c.109G>A (1/16). Additional GJB2 cases demonstrating a compound heterozygous state (5/16) were c.[35delG]; [139G>T] (2/16); c.[35delG]; [c.313_326del] (2/16), and c.[35delG]; [c.269T>C] (1/16) (see Table in Supplemental Digital Content 3, http://links.lww.com/EANDH/A960).
Furthermore, screening for OTOF revealed one compound heterozygous case with two novel causative alleles (c.[2719C>T]; c.[5203C>T]) not described in DVD or ClinVar at the time of diagnosis. Since then, the c.[5203C>T] variant has been registered in the HGMD and was recently diagnosed as a biallelic OTOF mutation in a Taiwanese family that presented with a phenotype compatible with auditory neuropathy of autosomal-recessive inheritance (Wu et al. 2019). Screening of SLC26A4 revealed one case with two known alleles ([c.918+1G>T]; [c.1001+1G>A]), the latter of which was previously described in European populations (Coyle et al. 1998; Ladsous et al. 2014).
Targeted Hearing Loss Gene Panel (NGS)
Subsequent variant identification by targeted hearing loss gene panel sequencing in 284 cases solved 57 cases (20%; n = 57/284) (see Tables in Supplemental Digital Content 3, http://links.lww.com/EANDH/A960 and 4 http://links.lww.com/EANDH/A961). No variants were identified in 57 cases (20%). In the remaining 170 patients (60%), a genetic diagnosis could not be conclusively made according to current knowledge and ACMG/AMP criteria (Richards et al. 2015; Oza et al. 2018) (i.e., VUS or only one heterozygous P or LP variant that was identified in a gene with autosomal-recessive inheritance).
Discussion
Phenotypic Characteristics of the Cohort
The unique feature of the large German cohort in this study is its age structure, which is highly representative of the age structure of the regional population in Southwest Germany (Fig. 1A). More importantly, this age structure is also reflected in the distribution of hearing loss onset, with the majority of patients showing late hearing loss onset in the postlingual domain (Fig. 1B). This is unusual, as most cohorts used in comprehensive targeted genetic testing efforts for hereditary hearing loss have had early hearing loss onset with a predominance in the congenital and pre/perilingual domains (Sloan-Heggen et al. 2016; Sommen et al. 2016; Zazo Seco et al. 2017). This may have resulted in potential bias toward phenotypic characteristics such as autosomal-recessive inheritance, bilateral symmetric hearing loss, and higher grades of hearing loss and hence influenced the solve rates.
Relationship of Phenotype to Diagnostic Yield
The overall diagnostic yield of comprehensive targeted genetic testing in the present German hearing loss cohort of 305 unrelated probands was 25%. However, the solve rate was sensitive to phenotypic characteristics in the categories of inheritance, hearing loss onset, and hearing loss grade. Single characteristics, such as autosomal-recessive inheritance, early hearing loss onset, or a grade of severe-profound-complete hearing loss, significantly elevated the solve rate. When these phenotypic characteristics were combined, solve rates at or above 60% were reached (Fig. 3B). Conversely, postlingual hearing loss onset or lower grades of hearing loss led to a significant decline in the solve rate (Fig. 3A). As the majority of the individuals were of Western European ethnicity (79%) and other ethnicities showed no significant differences in the solve rate compared to the overall solve rate (Fig. 3A), it seems appropriate to compare these results to those for other Western European and Caucasian cohorts.
At first glance, the diagnostic rate of 25% of the present study appears similar to that of a Western European hearing-impaired cohort with a Belgian-German background with a diagnostic rate of 25% to 30%. The Belgian-German study included 131 patients with autosomal-recessive NSHL and combined prescreening of GJB2 with targeted panel sequencing of 79 hearing loss genes. Based on previous data, the contribution of GJB2 was estimated to be 15% to 20%, and the increase in sensitivity by targeted panel sequencing of non-GJB2 genes was assessed to be 10% for this Belgian-German cohort (Sommen et al. 2016). However, this increase in sensitivity included only P genotypes. By incorporating LP genotypes, an additional 12% increase in sensitivity would lead to an increase in sensitivity by panel sequencing of 22%, raising the overall estimated solve rate to 35% to 40%.
The contribution of GJB2 to the general diagnostic yield of the present study was only 6% (n = 19 of 305). The increase in sensitivity by targeted panel sequencing of non-GJB2 genes summed to 20% (n = 60 of 305), which is a 1:3 relation instead of a 1:1 relation found for GJB2 and non-GJB2 genes in the Belgian-German study. The discrepancies between the two studies can be explained in part by two specific phenotypic inclusion criteria of the Belgian-German study: prelingual hearing loss onset and autosomal-recessive mode of inheritance. Considering only patients with pre/perilingual (including congenital) hearing loss onset (early onset) in the present study (42%; n = 127 of 305), the contribution of GJB2 to the diagnostic yield rises to 13% (n = 17 of 127) (Figs. 1B and 5A and B). This is within the range of the estimated GJB2 contribution in the Belgian-German cohort of 15% to 20%. Considering the non-GJB2 genes, the increase in sensitivity in the early hearing loss onset group reaches 28% (n = 36 of 127; P and LP classifications), resulting in an overall solve rate of 43% (n = 54 of 127) for this group (Fig. 3A). This is slightly above the estimated rate for the Belgian-German cohort (35%–40%). However, only half of the probands with an early onset also had an autosomal-recessive mode of inheritance, while the other half were distributed among autosomal-dominant and sporadic modes of inheritance and one X-linked case. Combining early onset and autosomal-recessive inheritance, the solve rate increases to 56% (n = 20 of 36) (Fig. 3B) which is approximately 20% above the estimated diagnostic rate of the Belgian-German cohort applying with the same inclusion criteria. This higher solve rate may be due to further phenotypic variations such as hearing loss grade, laterality, and regional ethnicity variations, as well as the greater number of genes in the respective hearing loss gene panels (79 versus 71–179 in the present study).
A second Western European study with a Dutch background obtained a solve rate of 33.5% (n = 67 of 200 patients) using exome sequencing followed by targeted hearing loss gene panel analysis of 120 genes (Zazo Seco et al. 2017), which is 8.5% higher than the present study. Although the Dutch study included all ages of hearing loss onset and modes of inheritance, the cohort was dominated (70%) by hearing loss onset in the congenital domain (n = 79) and the first decade of life (n = 60), with an even higher combined solve rate for these two groups of 44% (n = 61 of 139) (Zazo Seco et al. 2017). In the present study, the proportion of the same age group (0–9 years) in the overall cohort (42.3%; n = 129 of 305) was much smaller and not dominant (42.3% versus 70% in the Dutch cohort) (Fig. 1B). However, considering absolute size (n = 129) and the specific solve rate (43%; n = 56 of 129), this group with early onset shows the same results as those in the Dutch study (44%; n = 61 of 139). Interestingly, for late hearing loss onset in all subsequent decades (≥10 years), the solve rate in the Dutch study declined to 10% (n = 6 of 61), which is close to that in the present study, with a solve rate of 12% (n = 22 of 178) for the same late onset group (≥10 years). (The numbers are slightly different from those in the postlingual analysis shown in Fig. 3A, as 2 probands from the hearing loss group with onset at 7–9 years were added to the first decade for exact comparison.) Although the absolute number of probands in the late onset group was almost three times higher (n = 178 versus 61) than that in the Dutch study, the solve rate remained relatively low, emphasizing the diagnostic gap for postlingual hearing loss onset.
One of the largest studies to date used a gene panel platform targeting 66 or 89 deafness-associated genes and enrolled 1119 patients with a more diverse ethnic background in the United States (University of Iowa), with no exclusion criteria based on phenotype or inheritance. This cohort exhibited a diagnostic yield of 39% (n = 440) contributed by 49 genes (Sloan-Heggen et al. 2016). This may seem to be a much higher rate than that (25%) obtained in the present study. However, the diagnostic rate in the Iowa study also varied considerably based on phenotypic characteristics (Sloan-Heggen et al. 2016). Approximately half of the probands had Caucasian ethnicity, allowing for comparisons with the abovementioned Western European studies. Similar to the result in the Dutch study (Zazo Seco et al. 2017), 85% (954 of 1119) of the probands had congenital (56%) or childhood (29%) hearing loss onset (Sloan-Heggen et al. 2016). Patients with congenital hearing loss had a significantly greater diagnostic rate (44%) than patients with childhood (29%) or adult (28%) hearing loss onset (Sloan-Heggen et al. 2016). Combining congenital and childhood onset in the Iowa cohort, the solve rate is calculated at 33%, which is actually lower than the early onset (0–9 years) solve rate of 43% in the present and 44% in the Dutch studies. The predominance of early onset in the Iowa study (85% versus 44% in the present study) essentially explains the difference in the overall solve rate of 39% versus the 25% observed here. A recent data update of the Iowa cohort with 2460 individuals confirmed an overall solve rate of 39.9%. For congenital and childhood (3–6 years) onset, the solve rates were noted at 53.6% and 39.6%, respectively (Shearer et al. 2019). This compares to almost equivalent solve rates for congenital and childhood (pre/perilingual) onset of 48% and 40%, respectively, in the present study and reflects similar age-related declines in the two cohorts.
In summary, the diagnostic rates in targeted genetic studies are highly sensitive to phenotypic characteristics. If phenotypes are carefully controlled, surprising accordance of diagnostic rates emerges between these studies.
Gene Distribution
All 35 genes that impacted the diagnostic yield of this study are listed by the HL GCEP (DiStefano et al. 2019). Accordingly, the clinical validity has been classified as “definitive” for 25 of these genes, as “moderate” for 1 gene, as “limited” for 1 gene and as “not yet assessed” or “not done” for 8 genes (see Table in Supplemental Digital Content 6, http://links.lww.com/EANDH/A963 showing the ClinGen HL GCEP classifications for all 35 genes that contributed to the diagnostic yield). The majority of the solved cases (79%; n = 59 of 75) were based on “definitive” genes, and the remaining cases were related to genes classified as “moderate” (1%; n = 1), “limited” (2%; n = 2), or “not done” (17%; n = 13) (see Table in Supplemental Digital Content 6, http://links.lww.com/EANDH/A963). The gene classification also revealed that 27% of the solved cases received a genetic diagnosis implicating a syndromic/NSHL mimic gene (see Table in Supplemental Digital Content 5, http://links.lww.com/EANDH/A962 showing cases in syndromic/NSHL mimic genes).
Nearly half of the genetic diagnoses were represented by only five frequently causative genes, namely, GJB2 (25%), MYO15A, WFS1, SLC26A4, and COL11A1 (all 5%). The relative distribution of the involved genes varied notably according to their known inheritance patterns (Fig. 4A–C) and hearing loss onset (Fig. 5A and B). Due to the unique age structure of the cohort representative of the local population (Fig. 1A), equal numbers of recessive (GJB2, MYO15A, and TMPRSS3) and dominant (WFS1, COL11A1, and COL4A3) genes were uncovered among the six most frequent causative genes. This is unusual and different from the observations in other cohorts that emphasize early hearing loss onset.
The autosomal-dominant mode of inheritance is generally expected to account for only 19% of prelingual NSHL cases, the X-linked mode is expected to account for <1%, and the autosomal-recessive mode is expected to account for 80% (Smith et al. 2005; Shearer et al. 2017), a pattern that is well supported by the results of the Iowa study, with 14% autosomal-dominant, 1% X-linked, and 85% autosomal-recessive genetic diagnoses (Sloan-Heggen et al. 2016). In the present study, the quota of autosomal-dominant genetic diagnoses was much higher, at 41% (n = 31). Likewise, autosomal-recessive cases represented only 56% (n = 42) of the genetic diagnoses, and X-linked cases represented 3% (n = 2). This unusually large proportion of autosomal-dominant genetic diagnoses most likely is related to the balanced age structure of this study representative of the local population and hence a much larger fraction of late onset hearing loss than in other cohorts. This high rate of autosomal-dominant genetic diagnoses is also reflected in the gene distribution as compared to that of other studies. For example, in the Iowa study, the six most frequent causal genes, GJB2 (21.6%), STRC (16.1%), SLC26A4 (6.6%), TECTA (5.2%), MYO15A (4.8%), and MYO7A (4.5%), which were involved in more than half of the diagnoses, are predominantly recessive genes (Sloan-Heggen et al. 2016). This includes autosomal-dominant modes of inheritance in genes that may exhibit an early onset, as in the case of TECTA (DFNA8/12) (Walls et al. 2020) and MYO7A (DFNA11) (Yamamoto et al. 2020), reflecting the overall predominance of early onset hearing loss in the Iowa cohort. The same six genes were confirmed as the most frequent causal genes by a recently enlarged data set of the same cohort with 2460 individuals (Shearer et al. 2019). The only two genes among the six most frequent causal genes that overlapped between the present study and the Iowa study were the genes GJB2 and MYO15A, both of which cause recessive NSHL.
In some populations, the prevalence of pathogenic variants in GJB2 has been assessed to account for up to 50% of the genetic diagnoses of NSHL (Denoyelle et al. 1999; Kenneson et al. 2002). A meta-analysis revealed an overall prevalence of biallelic GJB2-associated hereditary hearing loss of 17.3% worldwide, 27.1% for Europe, and 14.1% for Germany (Chan & Chang 2014). Applying broad inclusion criteria, including all types of unspecified hearing loss (including late onset), the global prevalence of GJB2 declined significantly to 12.3% (Chan & Chang 2014). In German cohorts, the prevalence of biallelic GJB2 variants associated with hereditary hearing loss shows a broad range. Over the last two decades, sequential studies have revealed diagnostic rates of 14.3% (Gabriel et al. 2001) (early hearing loss onset), 2.6% (Kupka et al. 2002) (early hearing loss onset), 16.7% (Zoll et al. 2003) (childhood and adult onset, not specified), 21% (Bartsch et al. 2010) (early hearing loss onset), 31% (Beck et al. 2015) (childhood onset), and 13.4% (Burke et al. 2016) (childhood and adult onset). The last study represents the largest cohort and a relatively broad age range. The diagnostic rate of 13.4% corresponds to the 14.1% estimate from the meta-analysis for Germany (Chan & Chang 2014; Burke et al. 2016). The present study reveals a much lower overall diagnostic yield for causative biallelic GJB2 variants of 6.2% (19 of 305). However, if calculated for early hearing loss onset, the prevalence increases to 14.2% (18 of 127), and for late onset, it declines to 0.6% (1 of 178). Interestingly, the overall prevalence of causative GJB2 mutations in the Caucasian ethnicity group of the Iowa study can be calculated at 8.2% (45 out of 549), which appears close to the overall value of 6.2% (19 out of 305) in the present study.
With regard to the diagnostic contribution among the solved cases, GJB2 accounted for 25% overall (19 of 75 solved cases). For the early onset group, the proportion increased to 33% and for the late onset group decreased to 5%. In the Iowa study, the diagnostic contribution of GJB2-related hearing loss to the solved cases in the Caucasian ethnicity group was 22% (corrected for GJB2 prescreening), which is close the present study. As the Iowa study was dominated (85%) by early hearing loss onset, the figures of the present study need to be narrowed down to this phenotype for comparison. With regard to early hearing loss onset and European ethnicity, the GJB2-related values for diagnostic contribution condense to 21.2% (14 of 66) in the present study. Applying the same phenotypic characteristics, the overall prevalence of the cohort slightly declines to 5.2% (14 of 269). Both values for diagnostic contribution and overall prevalence come remarkably close to the results from the Caucasian group in the Iowa cohort, 22% and 8.2%, respectively. Again, the example of GJB2 demonstrates that comparisons between cohorts need to be carefully tailored according to phenotypic characteristics before drawing conclusions.
The diagnostic contribution of the MYO15A (DFNB3) gene of 5% is close to those in the Dutch (5.9%) (Zazo Seco et al. 2017) and the Iowa (4.8%) (Sloan-Heggen et al. 2016) studies. Causative mutations in MYO15A are among the second most frequent cause of hearing loss in this study, including novel variants and medically significant reclassifications (Supplemental Digital Content 7, http://links.lww.com/EANDH/A964). In other cohorts, MYO15A was also ranked as one of the most frequent causes of NSHL (Rehman et al. 2016).
The third gene, TMPRSS3 (DFNB8/10), is known for its variable hearing loss onset. Depending on the type of mutation, two phenotypes have been described: a severe early onset prelingual type (DFNB10) and a milder late onset postlingual progressive type (DFNB8) (Weegerink et al. 2011). Most TMPRSS3 cases in this study had a postlingual onset, and this gene was the second most frequent causal gene in the postlingual domain (Fig. 5B).
All other genes involved in a frequent diagnosis of the cohort were primarily genes that cause autosomal-dominant hearing loss (WFS1, COL11A1, and COL4A3). The gene WFS1 (DFNA6/14/38) is among the 10 genes most commonly causing hearing loss in other cohorts (Shearer et al. 2019) and is recognized for its characteristic low-frequency-hearing-loss phenotype (Bespalova et al. 2001; Cryns et al. 2002). A recent study screening a large cohort of Japanese probands investigated distinct types of audiometric configurations in heterozygous WFS1 variants. Although low-frequency hearing loss proved to be the most prevalent type, some individuals also showed high-frequency hearing loss (Kobayashi et al. 2018). A case of heterozygous WFS1 autosomal-dominant hearing loss based on medically significant reclassification also demonstrated high-frequency hearing loss in this study (case 148; see Table in Supplemental Digital Content 7, http://links.lww.com/EANDH/A964 showing pedigree and audiogram).
Until recently, the gene COL11A1 was primarily associated with syndromic hearing loss manifested as autosomal-dominant Marshall syndrome (MRSHS), autosomal-dominant or autosomal-recessive Stickler syndrome type II (STL2) and autosomal-dominant or autosomal-recessive fibrochondrogenesis 1 (FBCG1) characterized by ophthalmologic, facial, and skeletal abnormalities (OMIM 2021b). Current evidence also identifies COL11A1 as a gene causing autosomal-dominant NSHL (DFNA37). The first reported case was caused by a heterozygous splice-site altering variant (c.652-2A>C) identified in a large European-American family, expanding the phenotypic spectrum of COL11A1 to NSHL (Booth et al. 2019). In the present study, two families with NSHL were identified, each with a causative splice-site altering variant (cases 262 and 273, see Table in Supplemental Digital Content 4, http://links.lww.com/EANDH/A961 showing the variants). The splice-site variant (c.652-1G>C) and a de novo variant (c.4338+2T>C) were further characterized and published while this article was under review. The splice-site variant (c.652-1G>C) affects the same intron 4 canonical splice site originally reported for the European-American family (c.652-2A>C) (Booth et al. 2019) but elicits a different splicing outcome (Rad et al. 2021). All other reported families also showed no phenotypic evidence of syndromic features, providing further confirmation of COL11A1 as an autosomal-dominant NSHL gene.
The COL4A3 gene is associated with syndromic hearing loss in autosomal-dominant Alport syndrome (OMIM 2021a). Two of four probands diagnosed with causal COL4A3 variants in this study were characterized by syndromic features, including a segregation-supported medically significant reclassification for the variant c.4882T>G previously classified as a VUS (case 32; see Table in Supplemental Digital Content 7, http://links.lww.com/EANDH/A964 showing pedigree and audiogram). The two other cases were nonsyndromic mimics. Hearing loss is a characteristic feature of Alport syndrome and may appear as the primary clinical manifestation. A genetic diagnosis may help identify early renal disease (Rosado et al. 2015).
Panel Version
When considering diagnostic yield in relation to the panel version employed (see Tables in Supplemental Digital Content 1, http://links.lww.com/EANDH/A958 and 2, http://links.lww.com/EANDH/A959 showing list of genes and transcripts, respectively), most of the patients were tested using hearing loss panels version 2 (35%; n = 100), containing 95 genes (recruitment period: May 2012–May 2013), and version 4 (45%; n = 127), containing 164 genes (recruitment period: December 2014–September 2017) (Table 3). Analysis of diagnostic yield per gene panel version showed a solve rate of 20.0% for version 2 and a solve rate of 20.5% for version 4 (Table 3). It is inferred that despite a stark increase of 69 genes (a growth of 73%) on gene panel version 4 compared to version 2, the solve rate increased by only 0.5%. This corresponds to the observation that almost all 35 genes that contributed to the 75 diagnoses were already represented in the early gene panel (version 2). This gene panel version contained 32 of 35 genes (91%) that contributed to 72 of 75 (96%) of the solved cases. Only one gene and two genes that were added to version 3 and version 4, respectively, contributed to the small increase in diagnostic yield. Two-thirds of the genes involved in a diagnosis (n = 22; 63%) accounted for only one diagnosis each.
TABLE 3.
Diagnostic yield according to gene panel version
| Panel Version | No. Genes | No. Probands | Solved Cases | Diagnostic Yield |
|---|---|---|---|---|
| Version 1 | 71 | 2 | 0 | 0% |
| Version 2 | 95 | 100 | 20 | 20.0% |
| Version 3 | 110 | 44 | 6 | 13.6% |
| Version 4 | 164 | 127 | 26 | 20.5% |
| Version 5 | 179 | 11 | 5 | 45.5% |
This observation agrees with updated results from the Iowa cohort reporting genetic testing data from 2460 hearing-impaired individuals that revealed diminishing returns for the use of a larger number of genes in hearing loss panel sequencing. In this recent data update of the Iowa cohort, 79 different genes were identified as causative, but half of the genes (n = 34; 43%) accounted for a low frequency of diagnoses, with only one or two cases per gene (Shearer et al. 2019). The results presented here are also in line with the notion that the continuous increase in the number of genes in targeted genetic screening is useful and should be maintained to identify rare diagnoses but may not significantly increase the diagnostic yield.
This is also confirmed by observations in hereditary retinal degeneration, another sensory disease, that was studied in a comprehensive cohort of patients diagnosed in the largest center for inherited retinal degeneration in Germany over a period of nine years (2010–2018). This retinal study on the genetics of retinal dystrophies applied similar methodologies in the same diagnostic laboratory (CeGaT), recruited through the same Medical Center (University of Tübingen Medical Center), was performed during the same study period (2010–2018) and had a similar geographic focus with regards to the catchment area of the probands in Southwest Germany. When diagnostic rates for the early retinal gene panel versions [version 1 (RET.1): 150 genes; version 2 (RET.2): 128 genes] were compared to the latest panel [version 9 (RET.9): 379 genes] only a marginal increase of the diagnostic rate (from 67.3% for RET.1 and RET.2 to 69.2% for RET.9) was detected (Weisschuh et al. 2020). This led to the similar conclusion that the genes with the highest disease-causing variant load had already been identified in the early retinal gene panel versions.
For hereditary hearing loss the observations mentioned above clearly shift the diagnostic bottleneck from the escalating addition of new genes that was eminent a decade ago (Hilgert et al. 2009) and was resolved by the introduction of targeted gene panels (Shearer et al. 2010) to the level of variant classification (Azaiez et al. 2018).
Variants of Unknown Significance
VUSs represented two-thirds (69%) of the variants in the analysis and hence were by far the largest group among the classified variants in this study. This dominance of VUSs is in accord with the VUS distribution of medically relevant hearing loss variants within coding and splice-site regions in the DVD (74.3%; 72,066 of 97,007) (Azaiez et al. 2018). In the present study, variants most frequently assigned to the VUS category were found in the large genes ADGRV1 (n = 21; 4.7%), CDH23 (n = 18; 4.1%), MYO7A (n = 16; 3.6%), and USH2A (n = 13; 2.9%). The correlation of larger gene size with a larger number of VUSs is expected and may correspond to the higher variant load of larger genes in the overall variant analysis of the DVD (Azaiez et al. 2018).
Future re-evaluation of the current data will improve diagnostic results over time as the understanding and interpretation of variants in hearing loss will improve. The development and refinement of databases such as ClinVar and the HGMD and disease-specific databases such as the DVD will support the interpretation of VUSs in the future (Azaiez et al. 2018). Moving from targeted panel sequencing or exome sequencing to whole-genome sequencing will reveal the noncoding pathogenic spectrum and further increase the complexity of the pursuit to integrate genomic and phenotypic data for clinical interpretation.
Copy Number Variations
CNVs are a major cause of NSHL and have been shown to contribute to between ≈18% (Shearer et al. 2014b) and 20% of genetic hearing loss diagnoses (Sloan-Heggen et al. 2016). In particular, CNVs of the STRC gene (DFNB16) (Verpy et al. 2001) provided nearly three of four causal CNVs (Shearer et al. 2014b) or 16.1% of the total diagnoses (Sloan-Heggen et al. 2016). In this study, we identified one patient with a homozygous STRC partial deletion (c.(?_3499)_(4993_?)del; see Table in Supplemental Digital Content 4, http://links.lww.com/EANDH/A961), representing only 1.3% of the total diagnoses. Furthermore, we identified four individuals with heterozygous deletions (see Table in Supplemental Digital Content 7, http://links.lww.com/EANDH/A964), corresponding to a carrier frequency of 1.8% (5/284), compared to 4.7% in a previous study (Shearer et al. 2014b). The gene-specific diagnostic yield and carrier frequency for CNVs in STRC was relatively low in the present study. This gene is frequently deleted in patients of European descent (Francey et al. 2012; Vona et al. 2015) and was determined by molecular genetic testing to be the second most implicated gene in patients of French, Czech, and German cohorts (Vona et al. 2015; Baux et al. 2017; Plevova et al. 2017). With respect to other CNVs, one heterozygous deletion of exons 32–37 was identified in the OTOG gene that was suspected to be in compound heterozygosity with a putative splice-altering variant (c.2796-3C>G). Of note, identification of CNVs based on sequence coverage depth as previously described (Dohrn et al. 2017) was introduced early in the course (November 2012) of this retrospective study and was consistently performed (with the exception of STRC in panel version 3, leaving out 44 of 305 probands; 14%) so that CNV analysis covered the majority of the probands.
Considering phenotypic characteristics, STRC has primarily been described as a major contributor to pediatric bilateral NSHL, especially in patients with mild-moderate hearing loss (Francey et al. 2012). Causal variants in STRC provided the most common diagnosis among patients with mild-to-moderate hearing loss in the Iowa cohort and accounted for 30% of diagnoses for this hearing loss grade. Mild-to-moderate hearing loss accounted for 44% and severe-to-profound hearing loss for 56% of the patient population in the Caucasian ethnicity group of the Iowa cohort (Sloan-Heggen et al. 2016). In the present study, the overall proportion of bilateral normal to mild-to-moderate hearing loss (<50 dB HL) was only 19%, which is less than half of the proportion in the Iowa cohort (Fig. 2D). Hence, the relatively small number of probands demonstrating the typical audiologic phenotypic characteristics for STRC may provide a possible explanation for the low diagnostic contribution of STRC-related CNVs.
Further expansion of the cohort will reveal whether the observed CNV frequency and STRC contribution remain representative of this population.
CONCLUSIONS
In summary, we present the first large study of comprehensive targeted hearing loss sequencing in a German cohort. The cohort is unique because of its age structure, which is representative of the overall age distribution in the regional population of Southwest Germany. Only six genes accounted for half of the diagnoses, and 35 genes contributed an overall diagnostic yield of 25%. The solve rate was highly sensitive to phenotypic characteristics. Gene-specific participation in the genetic diagnoses was strongly influenced by the large proportion of postlingual hearing loss onset, which was reflected in an unusually high—more than 40%—contribution of autosomal-dominant genes to the diagnoses. A sequential increase in gene content in the targeted panels did not equitably increase diagnostic returns. The diagnostic bottleneck appeared in the substantial number of identified VUSs. These VUSs hold promise for increasing the diagnostic rate in future re-evaluations based on a growing knowledge base.
ACKNOWLEDGMENTS
We thank the patients and their families for participating in our study, as well as the personnel at the Department of OHNS Tübingen, Praxis für Humangenetik Tübingen and CeGaT GmbH, Tübingen, Germany, for technical assistance. We thank Drs. Hannie Kremer, Nijmegen, and Susanne Kohl, Tübingen for comments on the revised manuscript. This publication is dedicated to the memory of our dear colleague and friend Dr. Marcus Müller, who unexpectedly died during the course of this project.
Supplementary Material
Footnotes
published online ahead of print November 8, 2021.
Saskia Dofek is currently at the Department of Otolaryngology—Head & Neck Surgery, Facial Plastic Surgery, St. Vincentius-Kliniken, Steinhäuserstraße 18, 76135 Karlsruhe, Germany. Barbara Vona is currently at the University Medical Center Göttingen Institute of Human Genetics and Institute for Auditory Neuroscience & InnerEarLab, Heinrich-Düker-Weg 12, 37073 Göttingen, Germany.
M.M. is deceased.
M.H., A.T., H.L., and S.B. conceived the study; B.S., S.F., M.S., F.B., and S.B. were involved in experimental design; P.G., T.S-M., B.S., M.S., S.F., H.L., and B.V. analyzed the sequencing data or were involved in data curation; A.T., M.M., H.L., and B.V. prepared the original draft; H.L. prepared the revised draft; A.T., M.M., H.L., and B.V. reviewed and edited the manuscript; M.M. and A.H. generated figures and performed statistical analysis; H.L., S.B., and A.T. were involved in supervision; A.T., H.L., P.G., T.S.-M., S.B., and M.H. revised the original manuscript and the revision. All authors—except M.M.—have read and approved the final revised manuscript. This research received no external funding.
B.S., S.F., M.S., F.B., and S.B. are current or former employees of Praxis für Humangenetik, Tübingen, and/or CeGaT GmbH, Tübingen. The employment relationship had no impact on data collection or interpretation or the writing of the manuscript. The other authors have no conflicts of interest to disclose.
Variants have been submitted to ClinVar under the accession IDs SCV001571715–SCV001572151 (www.ncbi.nlm.nih.gov/clinvar/).
Supplemental digital content is available for this article. Direct URL citations appear in the printed text and are provided in the HTML and text of this article on the journal’s Web site (www.ear-hearing.com).
REFERENCES
- Adzhubei I. A., Schmidt S., Peshkin L., Ramensky V. E., Gerasimova A., Bork P., Kondrashov A. S., Sunyaev S. R. (2010). A method and server for predicting damaging missense mutations. Nat Methods, 7, 248–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alford R. L., Arnos K. S., Fox M., Lin J. W., Palmer C. G., Pandya A., Rehm H. L., Robin N. H., Scott D. A., Yoshinaga-Itano C.; ACMG Working Group on Update of Genetics Evaluation Guidelines for the Etiologic Diagnosis of Congenital Hearing Loss; Professional Practice and Guidelines Committee. (2014). American College of Medical Genetics and Genomics guideline for the clinical evaluation and etiologic diagnosis of hearing loss. Genet Med, 16, 347–355. [DOI] [PubMed] [Google Scholar]
- Azaiez H., Booth K. T., Ephraim S. S., Crone B., Black-Ziegelbein E. A., Marini R. J., Shearer A. E., Sloan-Heggen C. M., Kolbe D., Casavant T., Schnieders M. J., Nishimura C., Braun T., Smith R. J. H. (2018). Genomic landscape and mutational signatures of deafness-associated genes. Am J Hum Genet, 103, 484–497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartsch O., Vatter A., Zechner U., Kohlschmidt N., Wetzig C., Baumgart A., Nospes S., Haaf T., Keilmann A. (2010). GJB2 mutations and genotype-phenotype correlation in 335 patients from germany with nonsyndromic sensorineural hearing loss: Evidence for additional recessive mutations not detected by current methods. Audiol Neurootol, 15, 375–382. [DOI] [PubMed] [Google Scholar]
- Baux D., Vaché C., Blanchet C., Willems M., Baudoin C., Moclyn M., Faugère V., Touraine R., Isidor B., Dupin-Deguine D., Nizon M., Vincent M., Mercier S., Calais C., García-García G., Azher Z., Lambert L., Perdomo-Trujillo Y., Giuliano F., Claustres M., et al. (2017). Combined genetic approaches yield a 48% diagnostic rate in a large cohort of French hearing-impaired patients. Sci Rep, 7, 16783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck C., Pérez-Álvarez J. C., Sigruener A., Haubner F., Seidler T., Aslanidis C., Strutz J., Schmitz G. (2015). Identification and genotype/phenotype correlation of mutations in a large German cohort with hearing loss. Eur Arch Otorhinolaryngol, 272, 2765–2776. [DOI] [PubMed] [Google Scholar]
- Bespalova I. N., Van Camp G., Bom S. J., Brown D. J., Cryns K., DeWan A. T., Erson A. E., Flothmann K., Kunst H. P., Kurnool P., Sivakumaran T. A., Cremers C. W., Leal S. M., Burmeister M., Lesperance M. M. (2001). Mutations in the Wolfram syndrome 1 gene (WFS1) are a common cause of low frequency sensorineural hearing loss. Hum Mol Genet, 10, 2501–2508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Booth K. T., Askew J. W., Talebizadeh Z., Huygen P. L. M., Eudy J., Kenyon J., Hoover D., Hildebrand M. S., Smith K. R., Bahlo M., Kimberling W. J., Smith R. J. H., Azaiez H., Smith S. D. (2019). Splice-altering variant in COL11A1 as a cause of nonsyndromic hearing loss DFNA37. Genet Med, 21, 948–954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boucher S., Tai F. W. J., Delmaghani S., Lelli A., Singh-Estivalet A., Dupont T., Niasme-Grare M., Michel V., Wolff N., Bahloul A., Bouyacoub Y., Bouccara D., Fraysse B., Deguine O., Collet L., Thai-Van H., Ionescu E., Kemeny J. L., Giraudet F., Lavieille J. P., et al. (2020). Ultrarare heterozygous pathogenic variants of genes causing dominant forms of early-onset deafness underlie severe presbycusis. Proc Natl Acad Sci U S A, 117, 31278–31289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunak S., Engelbrecht J., Knudsen S. (1991). Prediction of human mRNA donor and acceptor sites from the DNA sequence. J Mol Biol, 220, 49–65. [DOI] [PubMed] [Google Scholar]
- Burke W. F., Warnecke A., Schöner-Heinisch A., Lesinski-Schiedat A., Maier H., Lenarz T. (2016). Prevalence and audiological profiles of GJB2 mutations in a large collective of hearing impaired patients. Hear Res, 333, 77–86. [DOI] [PubMed] [Google Scholar]
- Celniker S. E., Wheeler D. A., Kronmiller B., Carlson J. W., Halpern A., Patel S., Adams M., Champe M., Dugan S. P., Frise E., Hodgson A., George R. A., Hoskins R. A., Laverty T., Muzny D. M., Nelson C. R., Pacleb J. M., Park S., Pfeiffer B. D., Richards S., et al. (2002). Finishing a whole-genome shotgun: Release 3 of the Drosophila melanogaster euchromatic genome sequence. Genome Biol, 3, RESEARCH0079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan D. K., & Chang K. W. (2014). GJB2-associated hearing loss: Systematic review of worldwide prevalence, genotype, and auditory phenotype. Laryngoscope, 124, E34–E53. [DOI] [PubMed] [Google Scholar]
- Collins F. S., Morgan M., Patrinos A. (2003). The Human Genome Project: Lessons from large-scale biology. Science, 300, 286–290. [DOI] [PubMed] [Google Scholar]
- Coyle B., Reardon W., Herbrick J. A., Tsui L. C., Gausden E., Lee J., Coffey R., Grueters A., Grossman4 A., Phelps P. D., Luxon L., Kendall-Taylor P., Scherer S. W., Trembath R. C. (1998). Molecular analysis of the PDS gene in Pendred syndrome. Hum Mol Genet, 7, 1105–1112. [DOI] [PubMed] [Google Scholar]
- Cryns K., Pfister M., Pennings R. J., Bom S. J., Flothmann K., Caethoven G., Kremer H., Schatteman I., Köln K. A., Tóth T., Kupka S., Blin N., Nürnberg P., Thiele H., van de Heyning P. H., Reardon W., Stephens D., Cremers C. W., Smith R. J., Van Camp G. (2002). Mutations in the WFS1 gene that cause low-frequency sensorineural hearing loss are small non-inactivating mutations. Hum Genet, 110, 389–394. [DOI] [PubMed] [Google Scholar]
- Cunningham L. L., & Tucci D. L. (2017). Hearing loss in adults. N Engl J Med, 377, 2465–2473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denoyelle F., Marlin S., Weil D., Moatti L., Chauvin P., Garabédian E. N., Petit C. (1999). Clinical features of the prevalent form of childhood deafness, DFNB1, due to a connexin-26 gene defect: Implications for genetic counselling. Lancet, 353, 1298–1303. [DOI] [PubMed] [Google Scholar]
- DiStefano M. T., Hemphill S. E., Oza A. M., Siegert R. K., Grant A. R., Hughes M. Y., Cushman B. J., Azaiez H., Booth K. T., Chapin A., Duzkale H., Matsunaga T., Shen J., Zhang W., Kenna M., Schimmenti L. A., Tekin M., Rehm H. L., Abou Tayoun A. N., Amr S.S.; ClinGen Hearing Loss Clinical Domain Working Group. (2019). ClinGen expert clinical validity curation of 164 hearing loss gene-disease pairs. Genet Med, 21, 2239–2247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dohrn M. F., Glöckle N., Mulahasanovic L., Heller C., Mohr J., Bauer C., Riesch E., Becker A., Battke F., Hörtnagel K., Hornemann T., Suriyanarayanan S., Blankenburg M., Schulz J. B., Claeys K. G., Gess B., Katona I., Ferbert A., Vittore D., Grimm A., et al. (2017). Frequent genes in rare diseases: Panel-based next generation sequencing to disclose causal mutations in hereditary neuropathies. J Neurochem, 143, 507–522. [DOI] [PubMed] [Google Scholar]
- Francey L. J., Conlin L. K., Kadesch H. E., Clark D., Berrodin D., Sun Y., Glessner J., Hakonarson H., Jalas C., Landau C., Spinner N. B., Kenna M., Sagi M., Rehm H. L., Krantz I. D. (2012). Genome-wide SNP genotyping identifies the Stereocilin (STRC) gene as a major contributor to pediatric bilateral sensorineural hearing impairment. Am J Med Genet A, 158A, 298–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuller C., Başkent D., Free R. (2019). Early deafened, late implanted cochlear implant users appreciate music more than and identify music as well as postlingual users. Front Neurosci, 13, 1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuller C., Mallinckrodt L., Maat B., Başkent D., Free R. (2013). Music and quality of life in early-deafened late-implanted adult cochlear implant users. Otol Neurotol, 34, 1041–1047. [DOI] [PubMed] [Google Scholar]
- Gabriel H., Kupsch P., Sudendey J., Winterhager E., Jahnke K., Lautermann J. (2001). Mutations in the connexin26/GJB2 gene are the most common event in non-syndromic hearing loss among the German population. Hum Mutat, 17, 521–522. [DOI] [PubMed] [Google Scholar]
- GBD 2015 Disease and Injury Incidence and Prevalence Collaborators. (2016). Global, regional, and national incidence, prevalence, and years lived with disability for 310 diseases and injuries, 1990-2015: A systematic analysis for the Global Burden of Disease Study 2015. Lancet, 388, 1545–1602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GBD Hearing Loss Collaborators. (2021). Hearing loss prevalence and years lived with disability, 1990-2019: Findings from the Global Burden of Disease Study 2019. Lancet, 397, 996–1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Global Burden of Disease Hearing Loss Expert Group. Stevens G., Flaxman S., Brunskill E., Mascarenhas M., Mathers C. D., Finucane M., et al. (2013). Global and regional hearing impairment prevalence: An analysis of 42 studies in 29 countries. Eur J Public Health, 23, 146–152. [DOI] [PubMed] [Google Scholar]
- Harrison S. M., Riggs E. R., Maglott D. R., Lee J. M., Azzariti D. R., Niehaus A., Ramos E. M., Martin C. L., Landrum M. J., Rehm H. L. (2016). Using ClinVar as a resource to support variant interpretation. Curr Protoc Hum Genet, 89, 8.16.1–8.16.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hebsgaard S. M., Korning P. G., Tolstrup N., Engelbrecht J., Rouzé P., Brunak S. (1996). Splice site prediction in Arabidopsis thaliana pre-mRNA by combining local and global sequence information. Nucleic Acids Res, 24, 3439–3452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilgert N., Smith R. J. H., Van Camp G. (2009). Forty-six genes causing nonsyndromic hearing impairment: Which ones should be analyzed in DNA diagnostics? Mutat Res, 681, 189–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- James S. L., Abate D., Abate K. H., Abay S. M., Abbafati C., Abbasi N., Abbastabar H., Abd-Allah F., Abdelalim J. A. A., Abdollahpour I., Abdulkader R. S., Abebe Z., Abera S. F., Abil O. Z., Abraha H. N., Abu-Raddad L. J., Abu-Rmeileh N. M. E., Accrombessi M. M. K., Acharya D., Acharya P., et al. (2018). Global, regional, and national incidence, prevalence, and years lived with disability for 354 diseases and injuries for 195 countries and territories, 1990-2017: A systematic analysis for the Global Burden of Disease Study 2017. Lancet, 392, 1789–1858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jian X., Boerwinkle E., Liu X. (2014). In silico prediction of splice-altering single nucleotide variants in the human genome. Nucleic Acids Res, 42, 13534–13544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karczewski K. J., Francioli L. C., Tiao G., Cummings B. B., Alföldi J., Wang Q., Collins R. L., Laricchia K. M., Ganna A., Birnbaum D. P., Gauthier L. D., Brand H., Solomonson M., Watts N. A., Rhodes D., Singer-Berk M., England E. M., Seaby E. G., Kosmicki J. A., Walters R. K., et al.; Genome Aggregation Database Consortium. (2020). The mutational constraint spectrum quantified from variation in 141,456 humans. Nature, 581, 434–443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelsell D. P., Dunlop J., Stevens H. P., Lench N. J., Liang J. N., Parry G., Mueller R. F., Leigh I. M. (1997). Connexin 26 mutations in hereditary non-syndromic sensorineural deafness. Nature, 387, 80–83. [DOI] [PubMed] [Google Scholar]
- Kenneson A., Van Naarden Braun K., Boyle C. (2002). GJB2 (connexin 26) variants and nonsyndromic sensorineural hearing loss: A HuGE review. Genet Med, 4, 258–274. [DOI] [PubMed] [Google Scholar]
- Kobayashi M., Miyagawa M., Nishio S. Y., Moteki H., Fujikawa T., Ohyama K., Sakaguchi H., Miyanohara I., Sugaya A., Naito Y., Morita S. Y., Kanda Y., Takahashi M., Ishikawa K., Nagano Y., Tono T., Oshikawa C., Kihara C., Takahashi H., Noguchi Y., et al. (2018). WFS1 mutation screening in a large series of Japanese hearing loss patients: Massively parallel DNA sequencing-based analysis. PLoS One, 13, e0193359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopanos C., Tsiolkas V., Kouris A., Chapple C. E., Albarca Aguilera M., Meyer R., Massouras A. (2019). VarSome: The human genomic variant search engine. Bioinformatics, 35, 1978–1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korver A. M., Smith R. J., Van Camp G., Schleiss M. R., Bitner-Glindzicz M. A., Lustig L. R., Usami S. I., Boudewyns A. N. (2017). Congenital hearing loss. Nat Rev Dis Primers, 3, 16094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar P., Henikoff S., Ng P. C. (2009). Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat Protoc, 4, 1073–1081. [DOI] [PubMed] [Google Scholar]
- Kupka S., Braun S., Aberle S., Haack B., Ebauer M., Zeissler U., Zenner H. P., Blin N., Pfister M. (2002). Frequencies of GJB2 mutations in German control individuals and patients showing sporadic non-syndromic hearing impairment. Hum Mutat, 20, 77–78. [DOI] [PubMed] [Google Scholar]
- Ladsous M., Vlaeminck-Guillem V., Dumur V., Vincent C., Dubrulle F., Dhaenens C. M., Wémeau J. L. (2014). Analysis of the thyroid phenotype in 42 patients with Pendred syndrome and nonsyndromic enlargement of the vestibular aqueduct. Thyroid, 24, 639–648. [DOI] [PubMed] [Google Scholar]
- Landrum M. J., Chitipiralla S., Brown G. R., Chen C., Gu B., Hart J., Hoffman D., Jang W., Kaur K., Liu C., Lyoshin V., Maddipatla Z., Maiti R., Mitchell J., O’Leary N., Riley G. R., Shi W., Zhou G., Schneider V., Maglott D., et al. (2020). ClinVar: Improvements to accessing data. Nucleic Acids Res, 48(D1), D835–D844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landrum M. J., Lee J. M., Benson M., Brown G., Chao C., Chitipiralla S., Gu B., Hart J., Hoffman D., Hoover J., Jang W., Katz K., Ovetsky M., Riley G., Sethi A., Tully R., Villamarin-Salomon R., Rubinstein W., Maglott D. R. (2016). ClinVar: Public archive of interpretations of clinically relevant variants. Nucleic Acids Res, 44(D1), D862–D868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., & Durbin R. (2010). Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics, 26, 589–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Löhler J., Walther L. E., Hansen F., Kapp P., Meerpohl J., Wollenberg B., Schönweiler R., Schmucker C. (2019). The prevalence of hearing loss and use of hearing aids among adults in Germany: A systematic review. Eur Arch Otorhinolaryngol, 276, 945–956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morton C. C., & Nance W. E. (2006). Newborn hearing screening–a silent revolution. N Engl J Med, 354, 2151–2164. [DOI] [PubMed] [Google Scholar]
- Olusanya B. O., Davis A. C., Hoffman H. J. (2019a). Hearing loss grades and the International classification of functioning, disability and health. Bull World Health Organ, 97, 725–728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olusanya B. O., Davis A. C., Hoffman H. J. (2019b). Hearing loss: Rising prevalence and impact. Bull World Health Organ, 97, 646–646A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- OMIM. (2021a). COLLAGEN, TYPE IV, ALPHA-3; COL4A3. (Contributors: Ada Hamosh - updated: July 10, 2015. Creation Date: Victor A. McKusick: 10/18/1988 ed.). Johns Hopkins University. [Google Scholar]
- OMIM. (2021b). COLLAGEN, TYPE XI, ALPHA-1; HGNC approved gene symbol: COL11A1. (Contributors: Cassandra L. Kniffin - updated: August 11, 2019. Creation Date: Victor A. McKusick: 9/29/1987 ed.). Johns Hopkins University. [Google Scholar]
- Oza A. M., DiStefano M. T., Hemphill S. E., Cushman B. J., Grant A. R., Siegert R. K., Shen J., Chapin A., Boczek N. J., Schimmenti L. A., Murry J. B., Hasadsri L., Nara K., Kenna M., Booth K. T., Azaiez H., Griffith A., Avraham K. B., Kremer H., Rehm H. L., et al.; ClinGen Hearing Loss Clinical Domain Working Group. (2018). Expert specification of the ACMG/AMP variant interpretation guidelines for genetic hearing loss. Hum Mutat, 39, 1593–1613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker M., & Bitner-Glindzicz M. (2015). Genetic investigations in childhood deafness. Arch Dis Child, 100, 271–278. [DOI] [PubMed] [Google Scholar]
- Plevova P., Paprskarova M., Tvrda P., Turska P., Slavkovsky R., Mrazkova E. (2017). STRC deletion is a frequent cause of slight to moderate congenital hearing impairment in the Czech Republic. Otol Neurotol, 38, e393–e400. [DOI] [PubMed] [Google Scholar]
- Rad A., Schade-Mann T., Gamerdinger P., Yanus G. A., Schulte B., Müller M., Imyanitov E. N., Biskup S., Löwenheim H., Tropitzsch A., Vona B. (2021). Aberrant COL11A1 splicing causes prelingual autosomal dominant nonsyndromic hearing loss in the DFNA37 locus. Hum Mutat, 42, 25–30. [DOI] [PubMed] [Google Scholar]
- Rehman A. U., Bird J. E., Faridi R., Shahzad M., Shah S., Lee K., Khan S. N., Imtiaz A., Ahmed Z. M., Riazuddin S., Santos-Cortez R. L., Ahmad W., Leal S. M., Riazuddin S., Friedman T. B. (2016). Mutational spectrum of MYO15A and the molecular mechanisms of DFNB3 human deafness. Hum Mutat, 37, 991–1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards S., Aziz N., Bale S., Bick D., Das S., Gastier-Foster J., Grody W. W., Hegde M., Lyon E., Spector E., Voelkerding K., Rehm H. L.; ACMG Laboratory Quality Assurance Committee. (2015). Standards and guidelines for the interpretation of sequence variants: A joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med, 17, 405–424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosado C., Bueno E., Fraile P., García-Cosmes P., González-Sarmiento R. (2015). A new mutation in the COL4A3 gene responsible for autosomal dominant Alport syndrome, which only generates hearing loss in some carriers. Eur J Med Genet, 58, 35–38. [DOI] [PubMed] [Google Scholar]
- Rubinstein W. S., Maglott D. R., Lee J. M., Kattman B. L., Malheiro A. J., Ovetsky M., Hem V., Gorelenkov V., Song G., Wallin C., Husain N., Chitipiralla S., Katz K. S., Hoffman D., Jang W., Johnson M., Karmanov F., Ukrainchik A., Denisenko M., Fomous C., et al. (2013). The NIH genetic testing registry: A new, centralized database of genetic tests to enable access to comprehensive information and improve transparency. Nucleic Acids Res, 41(Database issue), D925–D935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmucker C., Kapp P., Motschall E., Loehler J., Meerpohl J. J. (2019). Prevalence of hearing loss and use of hearing aids among children and adolescents in Germany: A systematic review. BMC Public Health, 19, 1277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma A., Dorman M. F., Spahr A. J. (2002). A sensitive period for the development of the central auditory system in children with cochlear implants: Implications for age of implantation. Ear Hear, 23, 532–539. [DOI] [PubMed] [Google Scholar]
- Shearer A. E., DeLuca A. P., Hildebrand M. S., Taylor K. R., Gurrola J., II, Scherer S., Scheetz T. E., Smith R. J. (2010). Comprehensive genetic testing for hereditary hearing loss using massively parallel sequencing. Proc Natl Acad Sci U S A, 107, 21104–21109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shearer A. E., Eppsteiner R. W., Booth K. T., Ephraim S. S., Gurrola J., II, Simpson A., Black-Ziegelbein E. A., Joshi S., Ravi H., Giuffre A. C., Happe S., Hildebrand M. S., Azaiez H., Bayazit Y. A., Erdal M. E., Lopez-Escamez J. A., Gazquez I., Tamayo M. L., Gelvez N. Y., Leal G. L., et al. (2014a). Utilizing ethnic-specific differences in minor allele frequency to recategorize reported pathogenic deafness variants. Am J Hum Genet, 95, 445–453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shearer A. E., Hildebrand M. S., Smith R. J. H. (2017). Hereditary hearing loss and deafness overview. In Adam M. P., Ardinger H. H., Pagon R. A. (Eds.), GeneReviews((R)) (pp. 1993–2021). University of Washington. [Google Scholar]
- Shearer A. E., Kolbe D. L., Azaiez H., Sloan C. M., Frees K. L., Weaver A. E., Clark E. T., Nishimura C. J., Black-Ziegelbein E. A., Smith R. J. (2014b). Copy number variants are a common cause of non-syndromic hearing loss. Genome Med, 6, 37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shearer A. E., Shen J., Amr S., Morton C. C., Smith R. J.; Newborn Hearing Screening Working Group of the National Coordinating Center for the Regional Genetics Networks. (2019). A proposal for comprehensive newborn hearing screening to improve identification of deaf and hard-of-hearing children. Genet Med, 21, 2614–2630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shearer A. E., & Smith R. J. (2012). Genetics: Advances in genetic testing for deafness. Curr Opin Pediatr, 24, 679–686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shearer A. E., & Smith R. J. (2015). Massively parallel sequencing for genetic diagnosis of hearing loss: The new standard of care. Otolaryngol Head Neck Surg, 153, 175–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sloan-Heggen C. M., Bierer A. O., Shearer A. E., Kolbe D. L., Nishimura C. J., Frees K. L., Ephraim S. S., Shibata S. B., Booth K. T., Campbell C. A., Ranum P. T., Weaver A. E., Black-Ziegelbein E. A., Wang D., Azaiez H., Smith R. J. H. (2016). Comprehensive genetic testing in the clinical evaluation of 1119 patients with hearing loss. Hum Genet, 135, 441–450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith R. J., Bale J. F., Jr, White K. R. (2005). Sensorineural hearing loss in children. Lancet, 365, 879–890. [DOI] [PubMed] [Google Scholar]
- Snoeckx R. L., Huygen P. L., Feldmann D., Marlin S., Denoyelle F., Waligora J., Mueller-Malesinska M., Pollak A., Ploski R., Murgia A., Orzan E., Castorina P., Ambrosetti U., Nowakowska-Szyrwinska E., Bal J., Wiszniewski W., Janecke A. R., Nekahm-Heis D., Seeman P., Bendova O., et al. (2005). GJB2 mutations and degree of hearing loss: A multicenter study. Am J Hum Genet, 77, 945–957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sommen M., Schrauwen I., Vandeweyer G., Boeckx N., Corneveaux J. J., van den Ende J., Boudewyns A., De Leenheer E., Janssens S., Claes K., Verstreken M., Strenzke N., Predöhl F., Wuyts W., Mortier G., Bitner-Glindzicz M., Moser T., Coucke P., Huentelman M. J., Van Camp G. (2016). DNA diagnostics of hereditary hearing loss: A targeted resequencing approach combined with a mutation classification system. Hum Mutat, 37, 812–819. [DOI] [PubMed] [Google Scholar]
- State Statistical Office Baden-Württemberg. (2021). Bevölkerung Baden-Württembergs nach Alters- und Geburtsjahren (bev_altersjahre.csv). Bevölkerung in Baden-Württemberg nach Migrationshintergrund im engeren Sinn (MZ-LA-Migr-LR-WE.csv). Durchschnittsalter der Bevölkerung nach Geschlecht (01035100.csv). https://www.statistik-bw.de/BevoelkGebiet.
- Statistics Division United Nations. (2021). Standard country or area codes for statistical use. Series M, No. 49 (M49 standard). https://unstats.un.org/unsd/methodology/m49/.
- Stenson P. D., Mort M., Ball E. V., Chapman M., Evans K., Azevedo L., Hayden M., Heywood S., Millar D. S., Phillips A. D., Cooper D. N. (2020). The Human Gene Mutation Database (HGMD®): Optimizing its use in a clinical diagnostic or research setting. Hum Genet, 139, 1197–1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens G., Flaxman S., Brunskill E., Mascarenhas M., Mathers C. D., Finucane M.; Global Burden of Disease Hearing Loss Expert Group. (2013). Global and regional hearing impairment prevalence: An analysis of 42 studies in 29 countries. Eur J Public Health, 23, 146–152. [DOI] [PubMed] [Google Scholar]
- Thorpe R. K., & Smith R. J. H. (2020). Future directions for screening and treatment in congenital hearing loss. Precis Clin Med, 3, 175–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tucci D. L., Wilson B. S., O’Donoghue G. M. (2017). The growing-and now Alarming-Burden of hearing loss worldwide. Otol Neurotol, 38, 1387–1388. [DOI] [PubMed] [Google Scholar]
- Tucker T., Marra M., Friedman J. M. (2009). Massively parallel sequencing: The next big thing in genetic medicine. Am J Hum Genet, 85, 142–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Camp G., & Smith R. J. H. (2021). Hereditary HEARING LOSS HOMEPAGE. https://hereditaryhearingloss.org.
- Van de Heyning P., Tavora-Vieira D., Mertens G., Rompaey V. V., Rajan G. P., Müller J., Hempel J. M., Leander D., Polterauer D., Marx M., Usami S. I., Kitoh R., Miyagawa M, Moteki H., Smilsky K., Baumgartner W. D., Keintzel T. G., Sprinzl G. M., Wolf-Magele A., Arndt S., et al. (2016). Towards a unified testing framework for single-sided deafness studies: A consensus paper. Audiol Neurootol, 21, 391–398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Eyken E., Van Camp G., Van Laer L. (2007a). The complexity of age-related hearing impairment: Contributing environmental and genetic factors. Audiol Neurootol, 12, 345–358. [DOI] [PubMed] [Google Scholar]
- Van Eyken E., Van Laer L., Fransen E., Topsakal V., Hendrickx J. J., Demeester K., Van de Heyning P., Mäki-Torkko E., Hannula S., Sorri M., Jensen M., Parving A., Bille M., Baur M., Pfister M., Bonaconsa A., Mazzoli M., Orzan E., Espeso A., Stephens D., et al. (2007b). The contribution of GJB2 (Connexin 26) 35delG to age-related hearing impairment and noise-induced hearing loss. Otol Neurotol, 28, 970–975. [DOI] [PubMed] [Google Scholar]
- Verpy E., Masmoudi S., Zwaenepoel I., Leibovici M., Hutchin T. P., Del Castillo I., Nouaille S., Blanchard S., Lainé S., Popot J. L., Moreno F., Mueller R. F., Petit C. (2001). Mutations in a new gene encoding a protein of the hair bundle cause non-syndromic deafness at the DFNB16 locus. Nat Genet, 29, 345–349. [DOI] [PubMed] [Google Scholar]
- von Gablenz P., Hoffmann E., Holube I. (2017). Prevalence of hearing loss in Northern and Southern Germany. HNO, 65(Suppl 2), 130–135. [DOI] [PubMed] [Google Scholar]
- von Gablenz P., Hoffmann E., Holube I. (2020). Gender-specific hearing loss in German adults aged 18 to 84 years compared to US-American and current European studies. PLoS One, 15, e0231632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vona B., Hofrichter M. A., Neuner C., Schröder J., Gehrig A., Hennermann J. B., Kraus F., Shehata-Dieler W., Klopocki E., Nanda I., Haaf T. (2015). DFNB16 is a frequent cause of congenital hearing impairment: Implementation of STRC mutation analysis in routine diagnostics. Clin Genet, 87, 49–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vona B., Müller M., Dofek S., Holderried M., Löwenheim H., Tropitzsch A. (2019). A big data perspective on the genomics of hearing loss. Laryngorhinootologie, 98(S 01), S32–S81. [DOI] [PubMed] [Google Scholar]
- Vona B., Müller T., Nanda I., Neuner C., Hofrichter M. A., Schröder J., Bartsch O., Läßig A., Keilmann A., Schraven S., Kraus F., Shehata-Dieler W., Haaf T. (2014). Targeted next-generation sequencing of deafness genes in hearing-impaired individuals uncovers informative mutations. Genet Med, 16, 945–953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vuckovic D., Mezzavilla M., Cocca M., Morgan A., Brumat M., Catamo E., Concas M. P., Biino G., Franzè A., Ambrosetti U., Pirastu M., Gasparini P., Girotto G. (2018). Whole-genome sequencing reveals new insights into age-related hearing loss: Cumulative effects, pleiotropy and the role of selection. Eur J Hum Genet, 26, 1167–1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walls W. D., Moteki H., Thomas T. R., Nishio S. Y., Yoshimura H., Iwasa Y., Frees K. L., Nishimura C. J., Azaiez H., Booth K. T., Marini R. J., Kolbe D. L., Weaver A. M., Schaefer A. M., Wang K., Braun T. A., Usami S. I., Barr-Gillespie P. G., Richardson G. P., Smith R. J., et al. (2020). A comparative analysis of genetic hearing loss phenotypes in European/American and Japanese populations. Hum Genet, 139, 1315–1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weegerink N. J., Schraders M., Oostrik J., Huygen P. L., Strom T. M., Granneman S., Pennings R. J., Venselaar H., Hoefsloot L. H., Elting M., Cremers C. W., Admiraal R. J., Kremer H., Kunst H. P. (2011). Genotype-phenotype correlation in DFNB8/10 families with TMPRSS3 mutations. J Assoc Res Otolaryngol, 12, 753–766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weisschuh N., Obermaier C. D., Battke F., Bernd A., Kuehlewein L., Nasser F., Zobor D., Zrenner E., Weber E., Wissinger B., Biskup S., Stingl K., Kohl S. (2020). Genetic architecture of inherited retinal degeneration in Germany: A large cohort study from a single diagnostic center over a 9-year period. Hum Mutat, 41, 1514–1527. [DOI] [PubMed] [Google Scholar]
- Wilson B. S., & Tucci D. L. (2021). Addressing the global burden of hearing loss. Lancet, 397, 945–947. [DOI] [PubMed] [Google Scholar]
- Wilson B. S., Tucci D. L., Merson M. H., O’Donoghue G. M. (2017). Global hearing health care: New findings and perspectives. Lancet, 390, 2503–2515. [DOI] [PubMed] [Google Scholar]
- World Health Organization. (2018). Addressing the Rising Prevalence of Hearing Loss. World Health Organization. [Google Scholar]
- World Health Organization. (2021). World Report on Hearing. World Health Organization. [Google Scholar]
- Wu C. C., Tsai C. Y., Lin Y. H., Chen P. Y., Lin P. H., Cheng Y. F., Wu C. M., Lin Y. H., Lee C. Y., Erdenechuluun J., Liu T. C., Chen P. L., Hsu C. J. (2019). Genetic epidemiology and clinical features of hereditary hearing impairment in the Taiwanese population. Genes (Basel), 10, E772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto N., Mutai H., Namba K., Goto F., Ogawa K., Matsunaga T. (2020). Clinical profiles of DFNA11 at diverse stages of development and aging in a large family identified by linkage analysis. Otol Neurotol, 41, e663–e673. [DOI] [PubMed] [Google Scholar]
- Zazo Seco C., Wesdorp M., Feenstra I., Pfundt R., Hehir-Kwa J. Y., Lelieveld S. H., Castelein S., Gilissen C., de Wijs I. J., Admiraal R. J., Pennings R. J., Kunst H. P., van de Kamp J. M., Tamminga S., Houweling A. C., Plomp A. S., Maas S. M., de Koning Gans P. A., Kant S. G., de Geus C. M., et al. (2017). The diagnostic yield of whole-exome sequencing targeting a gene panel for hearing impairment in The Netherlands. Eur J Hum Genet, 25, 308–314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoll B., Petersen L., Lange K., Gabriel P., Kiese-Himmel C., Rausch P., Berger J., Pasche B., Meins M., Gross M., Berger R., Kruse E., Kunz J., Sperling K., Laccone F. (2003). Evaluation of Cx26/GJB2 in German hearing impaired persons: Mutation spectrum and detection of disequilibrium between M34T (c.101T>C) and -493del10. Hum Mutat, 21, 98. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.