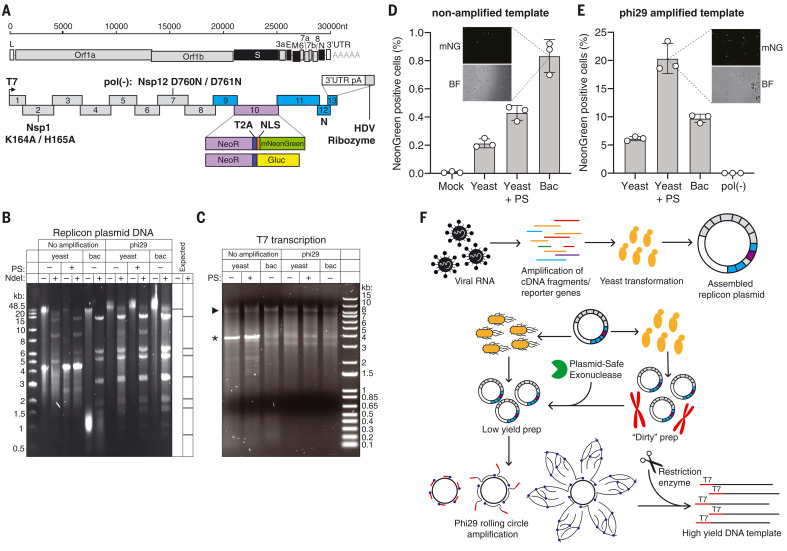
Fig. 1. SARS-CoV-2 replicon design and launch optimization.
(A) (Upper schematic) SARS-CoV-2 genome, with structural proteins in black. (Lower schematic) Replicon amplicon fragments for yeast assembly. Fragments from (10) are shown in gray; fragments harboring mutations in Nsp1 or Nsp12 [pol(-)] are marked as such. The reporter gene cassette in place of spike is shown in purple; reengineered flanking regions are in blue. nt, nucleotides; L, leader; UTR, untranslated region; pA: polyA, HDV, hepatitis delta virus; NLS, nuclear localization sequence. (B) Agarose gel of replicon DNA recovered from yeast or bacteria (bac). Phi29 amplification or plasmid-safe (PS) DNAse treatment is indicated. Expected NdeI digest is depicted at right. (C) Agarose gel of T7 RNA transcription reactions from the DNA plasmids in (B). The arrowhead indicates the expected size of full-length RNA; the asterisk denotes truncated product. (D and E) Percent of mNeonGreen replicon–positive BHK-21 cells from nonamplified (D) or phi29-amplified (E) DNA templates measured by flow cytometry. Insets show representative mNeonGreen (mNG) and bright-field (BF) images. N = 3 biological replicates. Error bars indicate SEM; “Mock” indicates no RNA electroporation. (F) Optimized RNA production for SARS-CoV-2 replicons. Overlapping PCR fragments are assembled in yeast and propagated in bacteria or yeast, in which case they are treated with PS DNAse. Subsequent phi29 amplification ensures full-length DNA template availability for transcription.