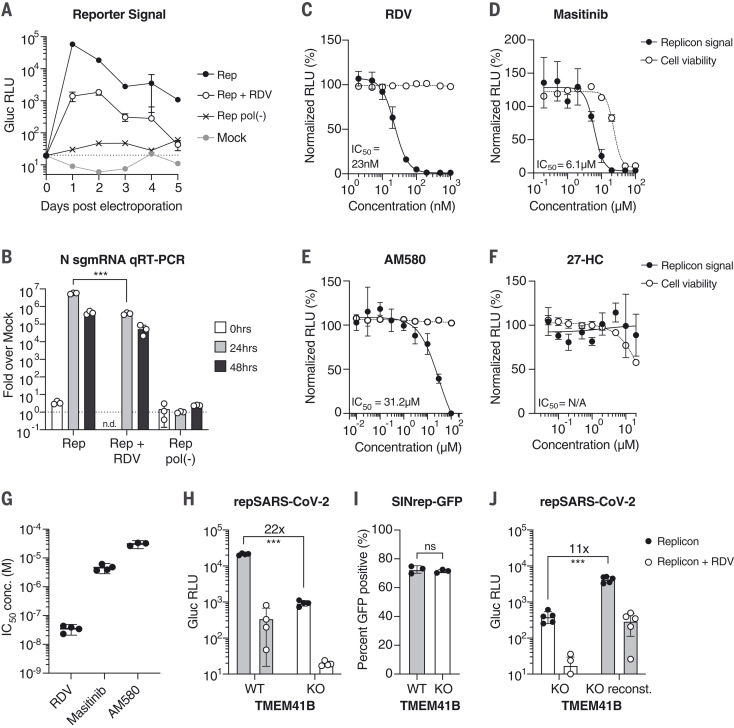
Fig. 2. SARS-CoV-2 replicons are sensitive to antiviral compounds, host factor loss, and viral mutant phenotypes.
(A) Gaussia luciferase (Gluc) in Huh-7.5 supernatant from cells electroporated with Gluc replicon RNA (Rep), seeded with 100 nM remdesivir (RDV) or vehicle. Mock electroporation and pol(-) replicons were used as controls. The dashed line indicates the limit of detection. N = 4. Error bars indicate SD. RLU, relative light units. (B) qRT-PCR measurements for subgenomic N RNA for cells in (A). Signal from mock-infected cells was used for normalization (dashed line). N = 3. Error bars indicate SD. ***P < 0.001; n.d., not determined. sgmRNA, subgenomic mRNA. (C to F) Representative experiments in Huh-7.5 cells were electroporated with the Gluc replicon RNA and seeded with (C) remdesivir (N = 4), (D) masitinib (N = 4), (E) AM580 (N = 3), or (F) 27-hydroxycholesterol (27-HC) (N = 3). After 24 hours, Gluc signal in the supernatant (filled circles) and cell viability (empty circles) were measured and normalized to vehicle-treated cells. Error bars indicate SEM. (G) IC50 values from independent experiments using the compounds presented in (C) to (E). (H) Parental Huh-7.5 (WT) and clonal TMEM41B KO cells were electroporated as indicated in (A). Gluc was measured 24 hours after electroporation. N = 4. (I) Cells as in (H) were electroporated with SINrep-GFP alphavirus replicon RNA. After 24 hours, GFP-positive cells were quantified by flow cytometry. N = 3. (J) As in (H), using cells reconstituted with TMEM41B. N = 5. Error bars indicate SD. ***P < 0.001 (two-sided Student’s t test); ns, not significant.