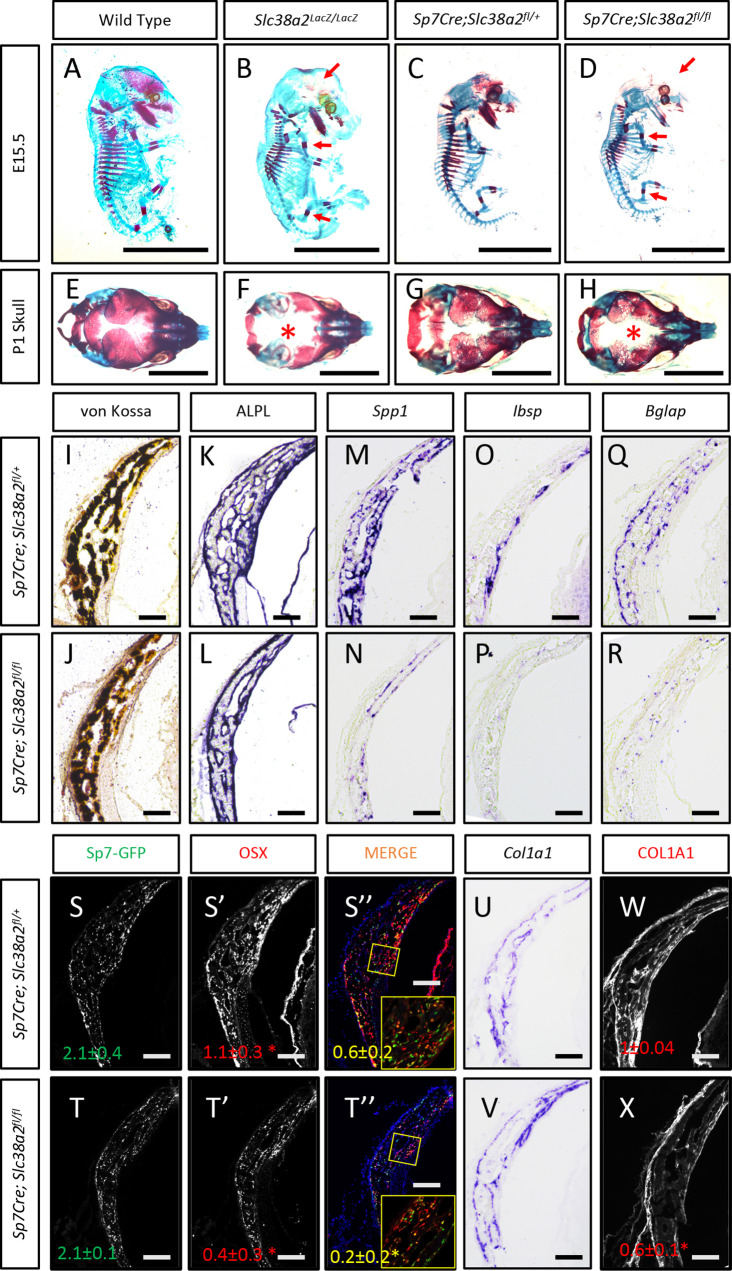
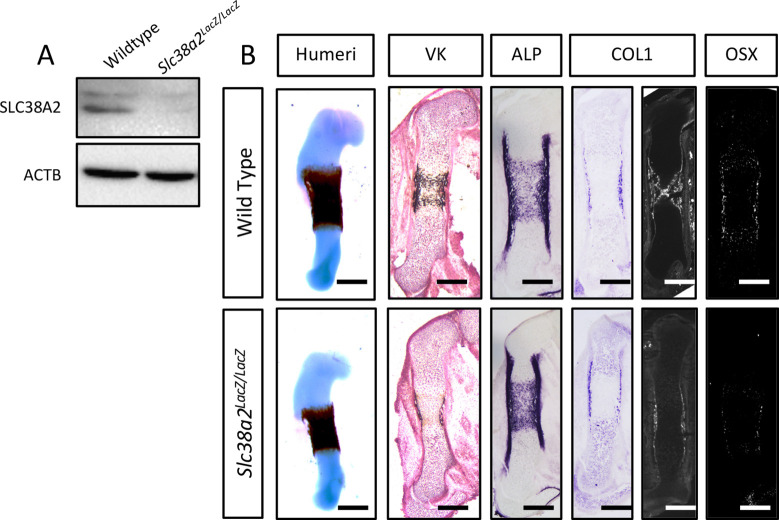
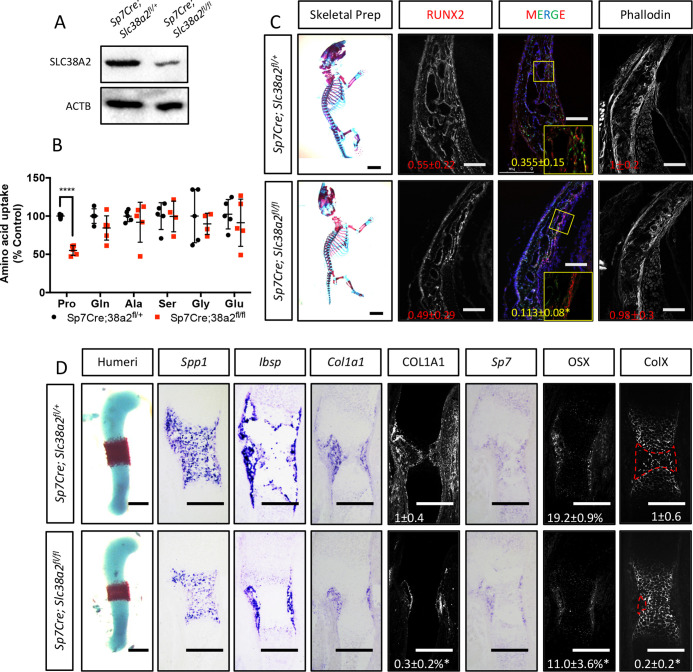
Figure 5. Slc38a2-dependent proline uptake is required for osteoblast differentiation during bone development.
(A–H) Skeletal preparations of Slc38a2LacZ/LacZ or wildtype controls (A, B, E, F) or Sp7Cre;Slc38a2fl/fl or Sp7Cre;Slc38a2fl/+ littermate controls (C, D, G, H) at embryonic day (E)15.5 (A–D) or P1 (E–H). Red arrow (A–D) or asterix (E–H) highlights reduced mineralization. A total of n = 7 or n = 5 Slc38a2LacZ/LacZ animals and n = 5 or n = 5 for Sp7Cre;Slc38a2fl/fl animals were analyzed at E15.5 or postnatal day (P)1, respectively. Scale bar = 5 mm. (I–R) Representative von Kossa staining (I, J), alkaline phosphatase (ALPL) staining (K, L) in situ hybridization for Spp1 (M, N), Ibsp (O, P), Bglap (Q, R), and Col1a1 (U, V), or immunofluorescent staining for OSX (S’, S”, T’, T”) and COL1A1 (W, X) on Sp7Cre;Slc38a2fl/fl (n = 4) (J, L, N, P, R, T, V, X) or Sp7Cre;Slc38a2fl/+ (n = 4) (I, K, M, O, Q, S, U, W) newborn calvariae. *p≤0.05 by paired two-tailed Student’s t-test. Scale bar = 100 μm.
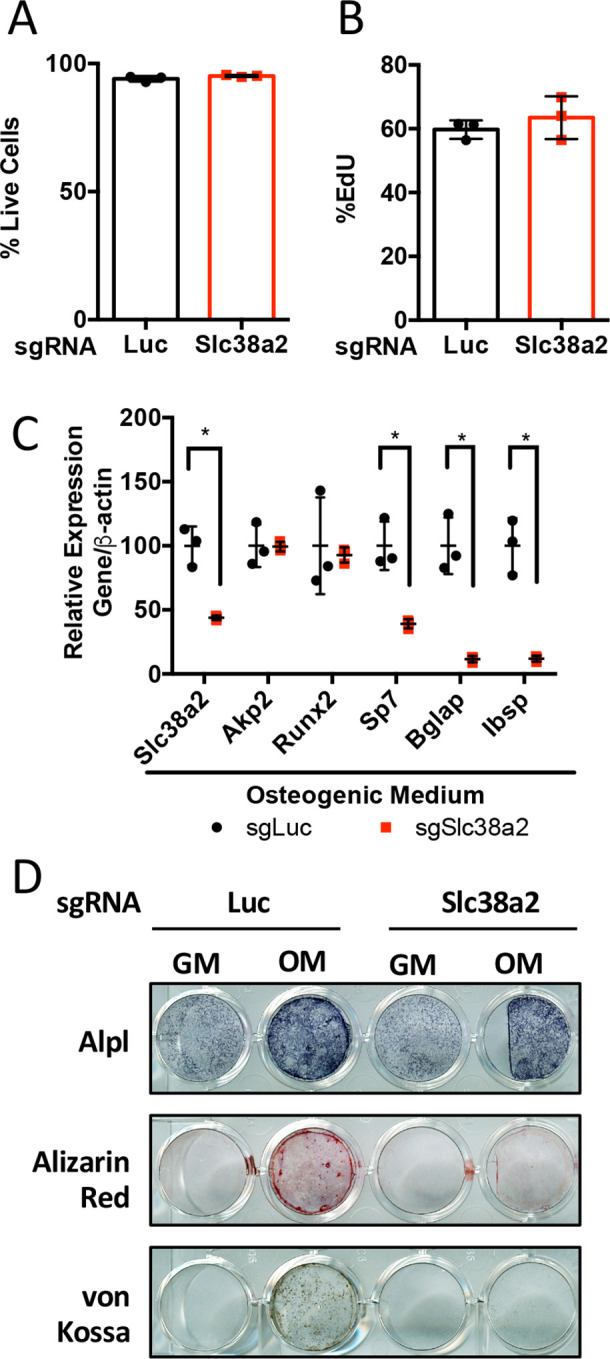
Figure 5—figure supplement 1. Slc38a2 is required for osteoblast differentiation in vitro.