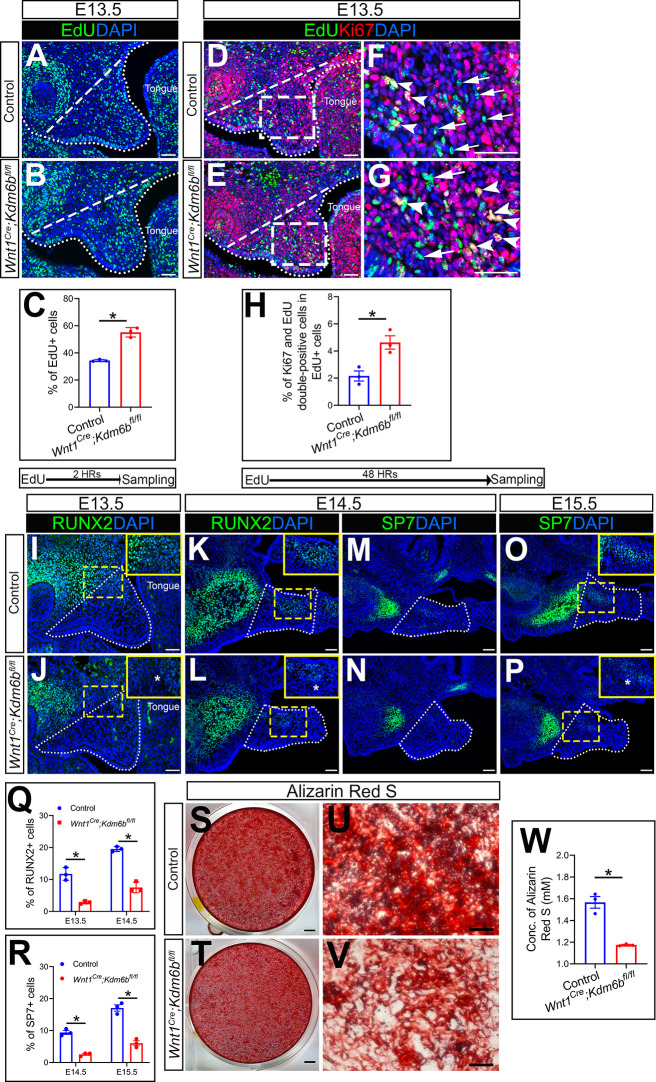
Figure 2. Kdm6b is critical for proliferation and differentiation of cranial neural crest (CNC)-derived palatal mesenchyme cells.
(A, B) Immunostaining of EdU at E13.5 after 2 hr of EdU labeling. Dotted lines indicate palatal shelf region. Dashed lines indicate the palatal region used for quantification in (C). Scale bar: 50 µm. (C) Quantification of EdU+ cells represented in (A, B). *p<0.05. (D–G) Co-localization of EdU and Ki67 at E13.5 after 48 hr of EdU labeling. Dotted lines indicate palatal shelf region. Dashed lines indicate the palatal region used for quantification in (H). (F, G) are magnified images of boxes in (D, E). Arrows in (F, G) indicate representative cells that are only EdU+, while arrowheads indicate representative cells that are positive for both EdU and Ki67. Scale bar: 50 µm. (H) Quantification of EdU and Ki67 double-positive cells represented in (D, E). *p<0.05. (I–L) Immunostaining of RUNX2 at indicated stages. Insets are higher-magnification images of boxes in (I–L). Asterisks in (J, L) indicate decreased RUNX2+ cells observed in Wnt1Cre;Kdm6bfl/fl mice. Scale bar: 50 µm. White dotted lines indicate the palatal region used for quantification in (Q). (M–P) Immunostaining of SP7 at indicated stages. Insets are higher-magnification images of boxes in (O, P). Asterisk in (P) indicates decreased SP7+ cells observed in Wnt1Cre;Kdm6bfl/fl mice. Scale bar: 50 µm. White dotted lines indicate the palatal region used for quantification in (R). (Q, R) Quantification results for RUNX2+ and SP7+ cells represented in (I–P). *p<0.05. (S–W) Osteogenic differentiation assay using Alizarin red S staining. (W) is the quantification result of Alizarin red S staining represented in (S, T). Scale bars: 2 mm in (S, T); 200 µm in (U, V). *p<0.05.