FIGURE 5.
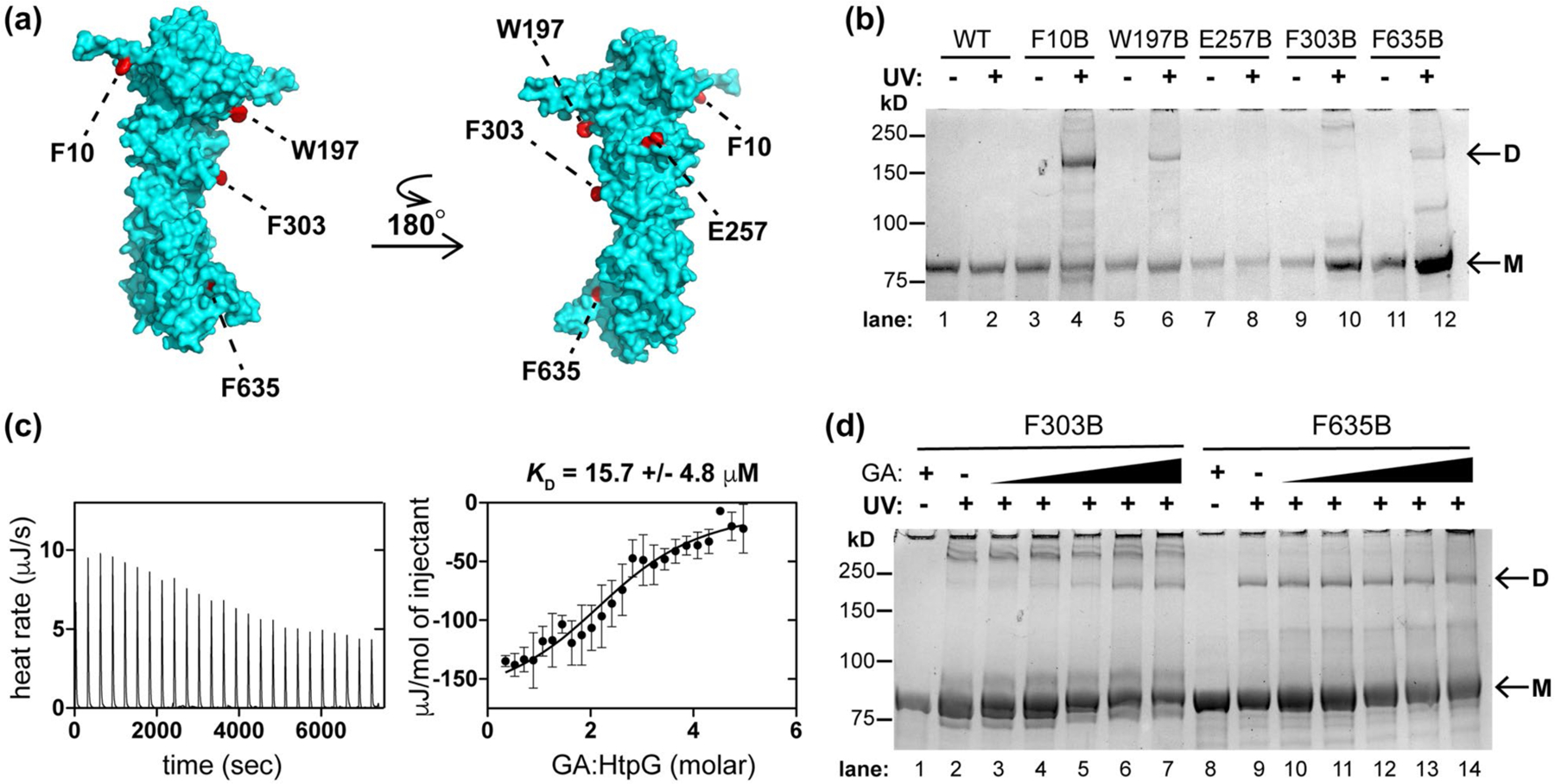
Mtb HtpG dimerizes in vitro and interacts with Hsp90 probe geldanamycin (GA). (a) Model of Mtb HtpG (Phyre2) (Kelley et al., 2015) with relevant residues highlighted that are sites of point mutation to the UV-activatable crosslinking amino acid para-benzoyl phenyalanine (BpF or B). (b) SDS-PAGE analysis of Mtb HtpG BpF mutants at indicated residues with and without prior UV irradiation (monomer bands are indicated as “M” and dimer bands are indicated as “D”). F10, W197, and F635 consistently mediate the formation of dimer. (c) ITC analysis of Mtb HtpG (47 µM) with GA (raw data is shown on left, analysis of average of three replicates with SD shown on the right) illustrates µM affinity. (d) SDS-PAGE analysis of indicated Mtb HtpG BpF mutants without (plus 400 µM GA) and with UV irradiation (in the presence of 0, 10, 50, 100, 200, and 400 µM GA) suggests that F303 turns into the dimer interface upon binding GA, while F635 is oriented in the dimer interface regardless of ligand binding. At least three identical analyses were performed for each experiment shown
