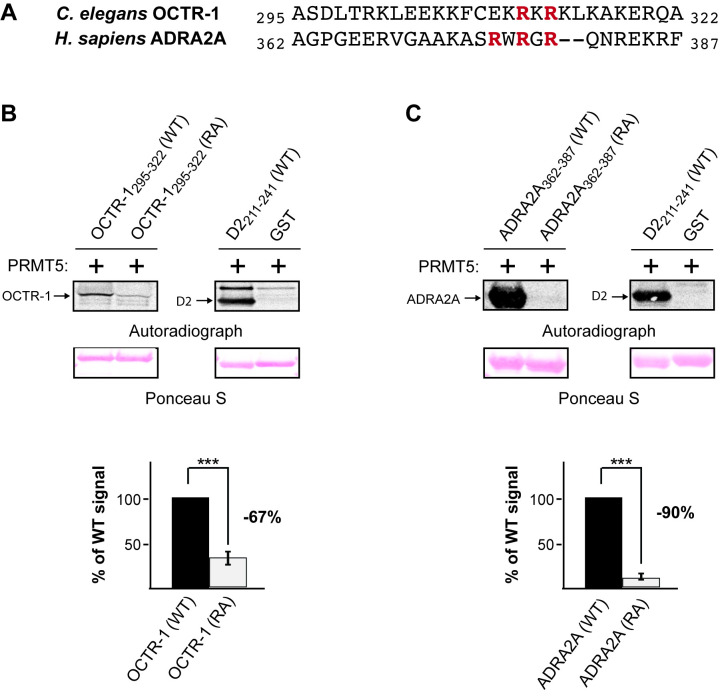
Figure 1. Human PRMT5 methylates the third intracellular loop of the C. elegans OCTR-1 receptor and the human alpha-2A adrenergic receptor (ADRA2A) in vitro .
(A) Alignment showing conservation of the predicted arginine methylation motifs in the C. elegans OCTR-1 and human ADRA2A receptors. The entire third intracellular loop (3 rd ICL) of OCTR-1 is predicted to have ~121 amino acids, representing residues 203-323 [SWISS-MODEL, based on (Qu et al. 2020)]; only residues 295-322 of the 3 rd ICL are shown and aligned with the corresponding residues of the 3 rd ICL of ADRA2A. Arginines within the predicted methylation motifs (RXR) are shown in red. (B and C) Representative blots for the in vitro methylation assays. A wild-type (WT) and mutant (RA) recombinant fragment of the 3 rd ICL of OCTR-1 (residues 295-322) and ADRA2A (residues 362-387), flanked both amino- and carboxy-terminally with an S-tag to increase solubility, and fused to glutathione S-transferase (GST) were used in an in vitro methylation assay with active recombinant human PRMT5. There are no arginines within the S-tag. A fragment of the human D2 dopamine receptor was used as the positive control for methylation (Likhite et al. 2015) and GST alone served as the negative control. Top: Autoradiographs show that WT OCTR-1 and ADRA2A are methylated by PRMT5. Fragments with the conserved arginines (shown in red, Figure 1A) changed to alanines were not efficiently methylated. Ponceau S staining of the polyvinylidene difluoride (PVDF) membranes was performed to demonstrate equivalent loading of the WT and RA receptor fragments. Bottom: Quantification of the degree of methylation of the receptor fragments based on densitometric analysis of the autoradiographs. Loss of the methylation motif resulted in a 67% decrease in OCTR-1 methylation and a 90% decrease in ADRA2A methylation. Data are means, +/- the standard error of the mean (SEM) from three independent experiments. *** = p < 0.001