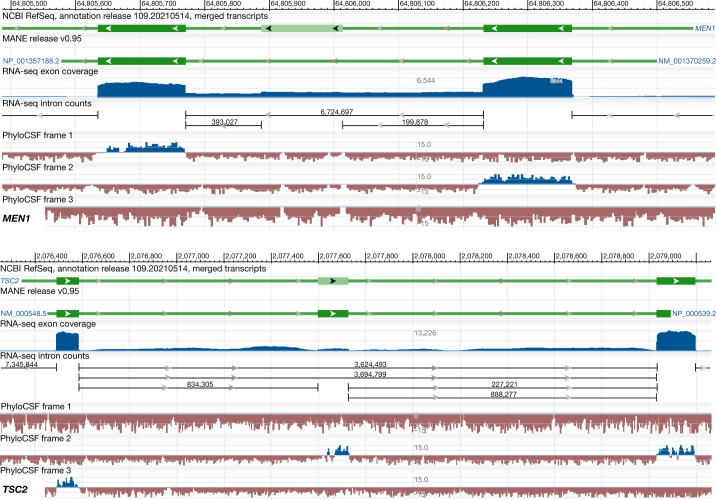
Fig. 1. Conservation versus expression when manually curating two high-value clinical genes.
Top, gene MEN1 (HGNC:7010) tracks from NCBI GDV, as described below from top to bottom. Track 1, magnified region of the gene showing a portion of the CDS including an alternatively spliced exon (NCBI annotation release 109.20210514). Track 2, MANE v0.95 track showing the corresponding region of the MANE Select transcript (NM_001370259.2) lacking the alternatively spliced exon. Track 3, RNA-seq exon coverage (aggregate, filtered), with the numbers indicating the peak heights of the graph on a linear scale. Track 4, RNA-seq intron-spanning data from recount3, with horizontal lines depicting introns and numbers above the line indicating the number of reads. Track 5, PhyloCSF tracks. A transcript excluding the alternatively spliced exon was chosen as the MANE Select transcript owing to low expression (tracks 3 and 4) and lack of evolutionary constraint (no positive PhyloCSF signal, as indicated by blue colour) for the alternatively spliced exon. Bottom, gene TSC2 (HGNC:12363) tracks from GDV, as described below from top to bottom. Track 1, NCBI annotation release 109.20210514 track showing a portion of the coding region. Track 2, MANE v0.95 track showing the corresponding region of the MANE Select transcript (NM_000548.5). Track 3, RNA-seq exon coverage (aggregate, filtered). Track 4, portion of RNA-seq intron-spanning data from recount3. Track 5, PhyloCSF tracks. The MANE Select transcript includes the alternatively spliced protein-coding exon, which, despite its lower expression compared with neighbouring exons, shows evolutionary constraint of the CDS (presence of positive signal in the PhyloCSF track, as indicated by blue colour).