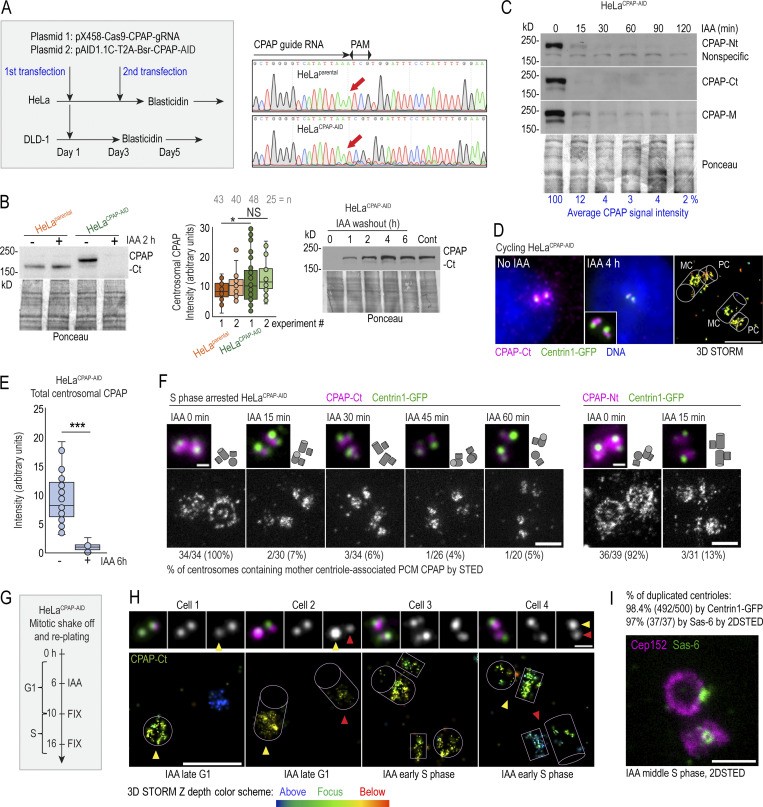
Figure 2.
Fast degradation of CPAP fused to an AID. (A) Left: The transfection and selection strategy (described in Materials and methods) used to generate HeLa and DLD-1 cell lines expressing CPAP fused to an AID (HeLaCPAP-AID and DLD-1CPAP-AID). Right: Sequencing result confirming the inactivation of CPAP alleles in HeLaCPAP-AID cells by a point mutation (red arrows). (B) Left and middle: Immunoblot and quantification of centrosomal CPAP in parental and HeLaCPAP-AID cells with and without IAA treatment. n = centrosome number. Right: Re-accumulation of CPAP-AID after IAA washout by immunoblot. (C) The efficiency and dynamics of CPAP-AID degradation after IAA addition. Antibodies recognizing Ct, Nt, and the middle (M) region of CPAP were used for detection. The residual levels of CPAP (shown in blue) represent an average signal intensity from the above blots. (D) Centrosomal CPAP levels and localization after 4 h of IAA treatment. Centrosome-associated CPAP is reduced, and residual CPAP signal is localized to centriole’s lumina and is absent from PCM. (E) Quantification of centrosomal CPAP from wide-field images before and after 6 h of IAA treatment. (F) 2DSTED analysis of centrosome-associated CPAP within first 60 min of IAA treatment. The wide-field images of corresponding superresolved centrioles are shown in insets. Centriole schemes depicting approximate centriole orientation are included. (G) Experimental strategy used in H and I. (H) 3DSTORM analysis of mother centrioles (MC) in late G1 (left) and in early S (right) in IAA-treated cells. CPAP signals analyzed by 3DSTORM are indicated by yellow and red arrows. (I) Centrosomes were labeled for mother centriole–specific protein Cep152 and procentriole-specific protein Sas-6. The percentage of duplicated mother centrioles was determined by scoring the number of MC-PC Centrin1-GFP pairs, and additionally by the presence of Sas-6 signal in association with Cep152 ring from 2DSTED recordings. Scale bars: 5 µm (wide-field); 0.5 µm (2DSTED and 3DSTORM); 1 µm (cropped centrosomes in F and H). *, P ≤ 0.05; ***, P ≤ 0.001.