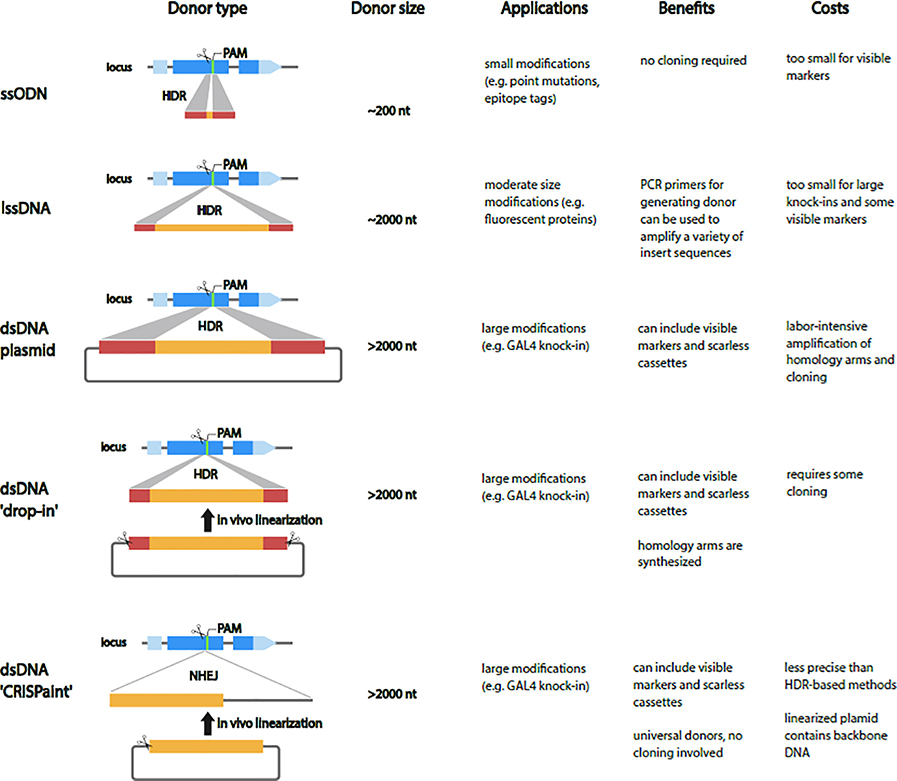
Figure 3. Types of Donor DNA for repair templates.
For small modifications short single-strand oligodeoxynucleotide (ssODN) repair templates are cheap and fast but are limited to a few hundred nucleotides. In the PCR-based long ssDNA (lssDNA) synthesis method, short homology arms are incorporated into PCR primers which amplify from a plasmid template that contains the insert sequence. The PCR product is treated with an exonuclease, which degrades one of the strands leaving behind a lssDNA homology donor. For large modifications, the most-used donors are double-strand DNA (dsDNA) donors supplied as circular plasmids. The main drawback of standard dsDNA donors is that they are labor intensive to generate and require long homology arms (0.5–1.5 kb) for efficient editing. The ‘drop-in’ strategy in which a plasmid containing a full-length dsDNA cassette with short homology arms is linearized in vivo combines the speed and cost effectiveness of ssODNs with the ability to include large repair templates. An alternative to the HDR knock-ins in Drosophila is the ‘CRISPaint’ method, which involves linearization of a universal circular donor plasmid, and integration into the target site by the NHEJ repair pathway.