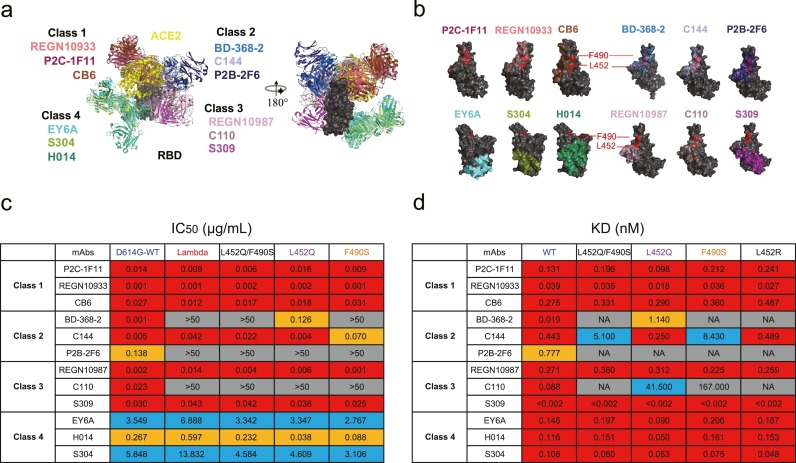
Fig. 2.
Neutralization of SARS-CoV-2 Lambda variant by monoclonal nAbs. (a) Structural depiction of ACE2 and representative nAbs from each class binding to the RBD. Class 1: P2C-1F11 (PDB: 7CDI), REGN10933 (PDB: 6XDG), CB6 (PDB: 7C01); Class 2: BD-368–2 (PDB: 7CHH), C144 (PDB: 7K90), P2B-2F6 (PDB: 7BWJ); Class 3: REGN10987 (PDB: 6XDG), C110 (PDB: 7K8V), S309 (PDB: 6WPS); Class 4: EY6A (PDB: 6ZCZ), H014 (PDB: 7CAI), S304 (PDB: 7JW0)[7–9]. (b) Footprints of four classes of representative nAbs on the RBD. L452 and F490 residuals are shown in red. (c) The IC50 values of nAbs against SARS-CoV-2 pseudoviruses. The data shown here are means of two independent experiments. The IC50 values below 0.05 μg/mL are marked in red. The IC50 values between 0.05 and 1 μg/mL are marked in orange. The IC50 values between 1 and 50 μg/mL are marked in blue. The IC50 values above 50 μg/mL are highlighted in gray. (d) The affinity values in binding activities of nAbs to mutated and WT RBD. The data shown here are means of two independent experiments. The binding affinities of nAbs below 0.5 nM are marked in red. The binding affinities of nAbs between 0.5 and 5 nM are marked in orange. The binding affinities of nAbs between 5 and 50 nM are marked in blue. The binding affinity above 50 nM, or not available (NA), in gray, indicates that the affinity of nAbs to the mutated RBD was very weak.