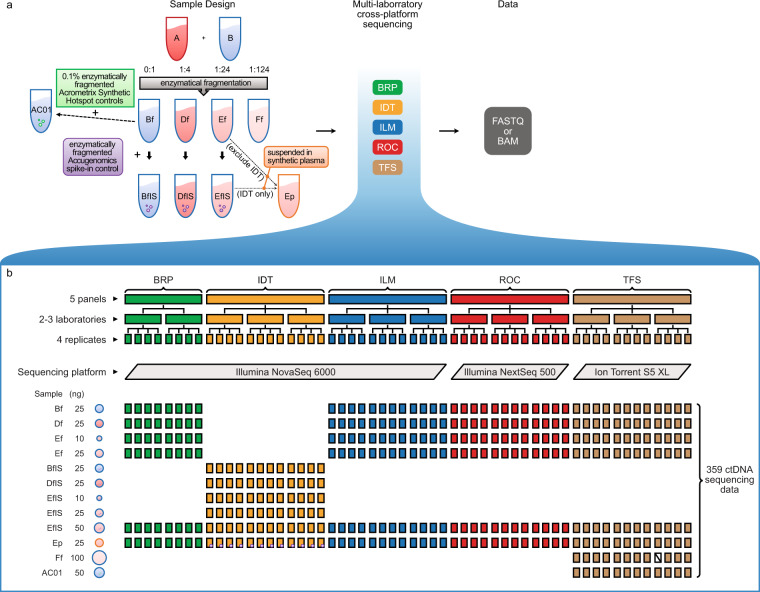
Fig. 1.
Study design. (a) Sample A and Sample B were mixed at ratios of 0:1, 1:4, 1:24, and 1:124, and then enzymatically fragmented to create Bf, Df, Ef, and Ff. Enzymatically fragmented Accugenomics spike-in control was added to Bf, Df, and Ef to create BfIS, DfIS, and EfIS. Sample Ef was suspended in synthetic plasma to create Ep. 0.1% enzymatically fragmented Acrometrix Synthetic Hotspot controls were added to Sample Bf to create Sample AC01. Sample Ef (Sample EfIS for panel IDT) was suspended in synthetic plasma solutions to create Sample Ep. (b) The illustration demonstrates the workflow of sample distribution, library preparation, and sequencing data processing. A set of reference samples were distributed to each of the testing laboratories. Four technical library replicates were prepared for each sample at each laboratory. DNA libraries were then sequenced using one of the three sequencing platforms. This figure is modified from Figure 3 in our related work4.