Figure 6.
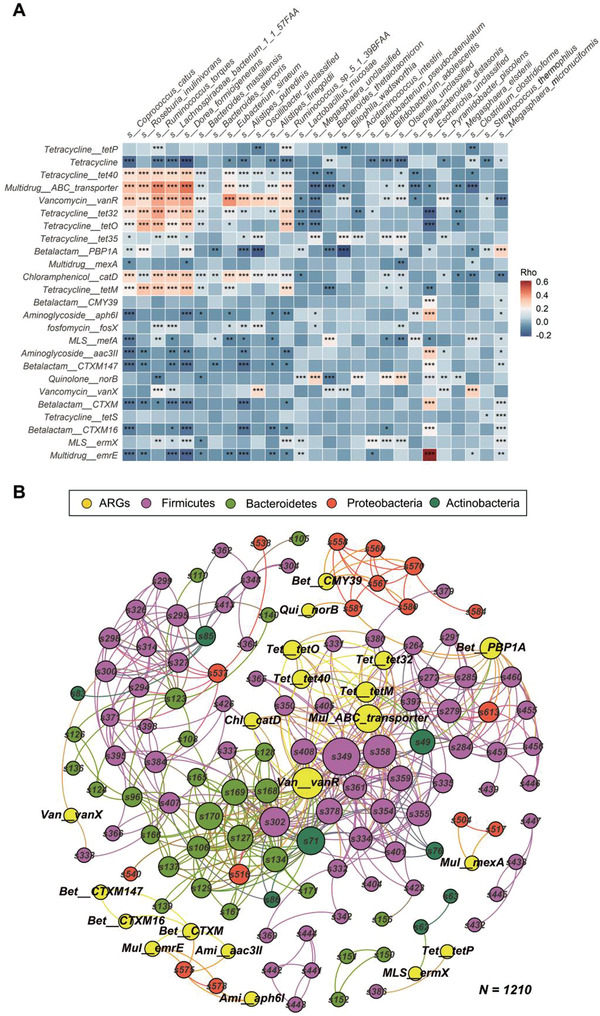
Co‐occurrence patterns among T2D‐related ARGs and microbial species. A) The heatmap shows the Spearman correlation coefficients between T2D‐related gut antibiotic resistance genes (ARGs) and T2D‐related microbial species. B) Correlation‐based networks of co‐occurring T2D‐related ARGs and all microbial species colored by node affiliation. A node stands for an ARG type/subtype or a species and a connection (i.e. edge) stands for a significant (FDR‐corrected p < 0.05, Spearman's rho ≥ 0.3, n = 1210) pairwise correlation. Node size is proportional to the number of connections (i.e., degree). Network was colored by ARGs and phylums. Ami, Aminoglycoside; Bet, Betalactam; Chl, Chloramphenicol; MLS, Macrolide‐Lincosamide‐Streptogramin; Mul, Multidrug; Qui, Quinolone; Tet, Tetracycline; Van, Vancomycin; *FDR‐corrected p < 0.05, ** FDR‐corrected p < 0.01, ***FDR‐corrected p < 0.001.
