Figure 6.
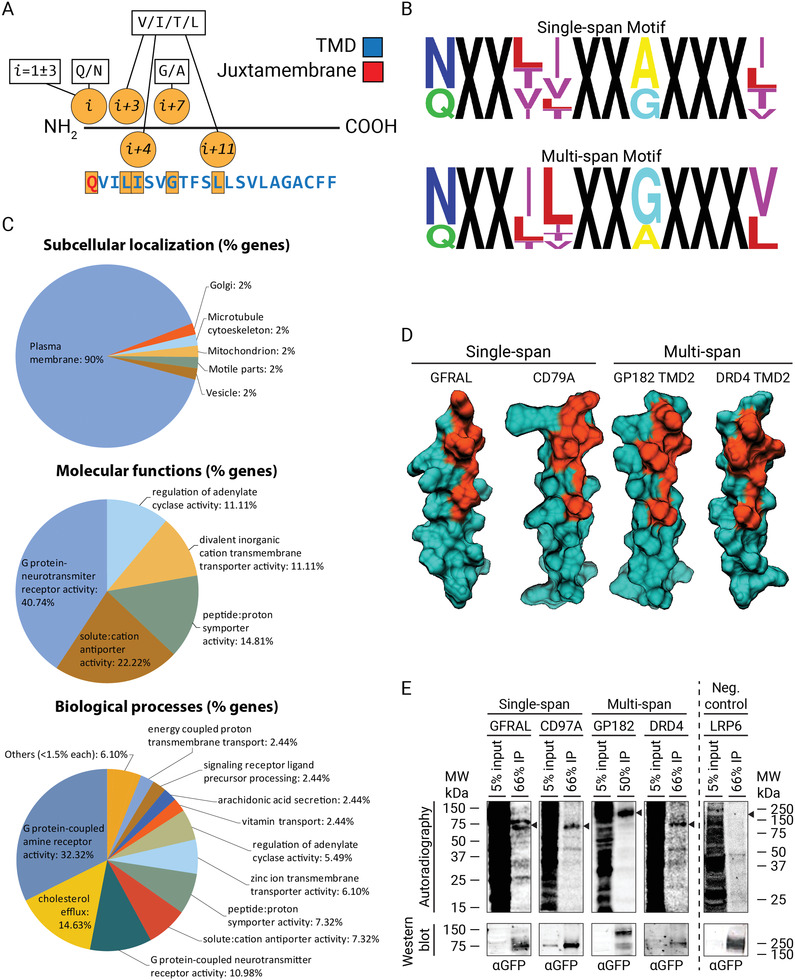
A general chol‐binding motif in mammalian membrane proteins. A) Relaxed motif signature used for bioinformatic studies using MOPRO for screening potential chol‐binding single‐ and multi‐span protein candidates in mammalian membrane proteome data sets. B) TMDs motifs significantly overrepresented with a p‐value ≤ 0.05 and z‐score > 2.5 were used to generate a single‐ (top panel) and multi‐span (bottom panel) sequence logo, where the letter size reflects the probability to find this amino acid at that particular position. C) Bioinformatic studies showing the subcellular distribution, molecular, and biological functions associated with the different putative chol‐binding membrane protein candidates identified in (A). D) 3D Minimal‐energy structures of the four candidates tested for chol‐binding in living cells. The IFN‐γR2 like chol‐binding domain is highlighted in orange, whereas the rest of the protein is depicted in dark green. E) In vivo labeling of GFP‐tagged constructs of candidates GFRAL, CD79A, GP182, DRD4, and the negative control LRP6. HAP1 cells were labeled with 100 μCi bifunctional Chol (3 µm) for 6 h, UV‐irradiated, lysed, and subjected to IP using an antibody against GFP. Corresponding radioactivity recovered for each protein candidate was detected and visualized by autoradiography and western blot analysis, respectively. Arrow, expected protein sizes (n = 3 independent experiments).
