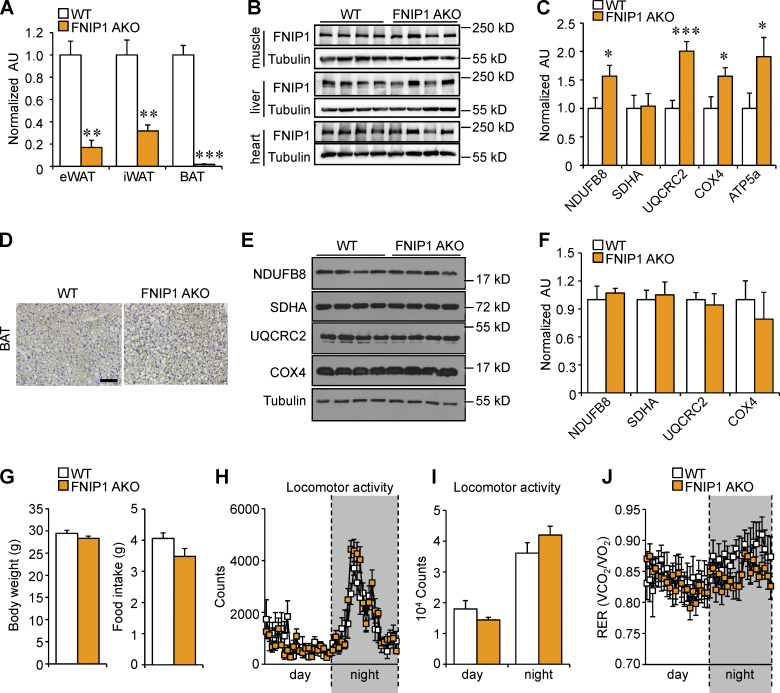
Figure S1.
Generation and characterization of mice with adipocyte-specific disruption of the Fnip1 gene. (A) qRT-PCR analysis of Fnip1 expression in eWAT, iWAT, and BAT of male WT and FNIP1 AKO mice. n = 3–6 mice per group. (B) Representative Western blot analysis of FNIP1 protein expression in skeletal muscle, liver, and heart from indicated male mice. n = 8 mice per group. (C) Quantification of the NDUFB8/tubulin, SDHA/tubulin, UQCRC2/tubulin, COX4/tubulin and ATP5A/tubulin signal ratios in Fig. 1 I were normalized (= 1.0) to the WT control mice. n = 11–12 mice per group. (D) IHC staining with a UCP1-specific antibody in BAT sections of indicated male mice. Scale bar: 20 μm. n = 8 mice per group. (E) Representative Western blot analysis of BAT lysates for male mice of the indicated genotypes using the indicated antibodies. (F) Quantification of the NDUFB8/tubulin, SDHA/tubulin, UQCRC2/tubulin, and COX4/tubulin signal ratios in E were normalized (= 1.0) to the WT controls. n = 4 mice per group. (G) Body weight of 12-wk-old male WT or FNIP1 AKO mice. n =8–12 mice per group. (H–J) Metabolic cage measurements of 16-wk-old male WT and FNIP1 AKO mice. n = 7–10 mice per group. Food consumption per day. (H and I) Locomotor activity over 12-h light/dark cycle. (J) Respiratory exchange rate (RER) during the light/dark cycle. Values represent mean ± SEM. *, P < 0.05; **, P < 0.01; ***, P < 0.001. P value was determined using two-tailed unpaired Student’s t-test (A, C, F, G, and H), or two-way ANOVA and Fisher’s LSD post-hoc test (H and J). Data are representative of at least two independent experiments (A–F). Source data are available for this figure: SourceData FS1.