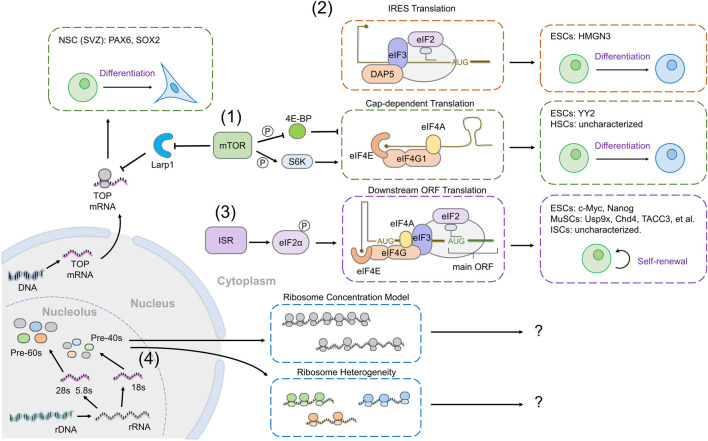
FIGURE 3.
Global translation mechanisms result in selective translation of specific transcripts to regulate stem cell fate. (1) Increased cap-dependent translation downstream of mTOR activity can result in selective translation of transcripts that have low translation efficiency. In ESCs, the mRNA encoding YY2 is one such target, in other tissues (HSCs), whether mTOR activity results in specific target expression is unknown (green box). mTOR also promotes the translation of mRNAs with TOP-like motifs, such as PAX6 and SOX2, which are translationally repressed during neuronal differentiation, in response to reduced mTOR activity. (2) Internal ribosome entry site (IRES)-mediated translation occurs when cap-binding is inhibited and can direct transcript-specific translation. In ESCs, DAP5 replaces eIF4G to promote IRES-dependent translation and ESC differentiation, through translation of HMGN3 (orange boxes). (3) p-eIF2ɑ selectively promotes the translation of mRNAs with an upstream open reading frame (uORF). In ESCs, p-eIF2ɑ promotes the translation of the mRNAs encoding c-Myc and Nanog. In MuSCs, p-eIF2ɑ promotes the translation of mRNAs coding for Usp9x, Chd4, TACC3, etc. (purple box). (4) Ribosomes can selectively regulate mRNA translation. Two models have been proposed, the ribosome concentration model in which the amount of ribosomes available affects the translation of transcripts with high or low translation efficiency differently, or ribosome heterogeneity in which different ribosomal subunit composition directs specific translation of particular transcripts (blue boxes). Although ribosomes impact stem cell fate, no evidence directly supports either model to date.