Figure 3.
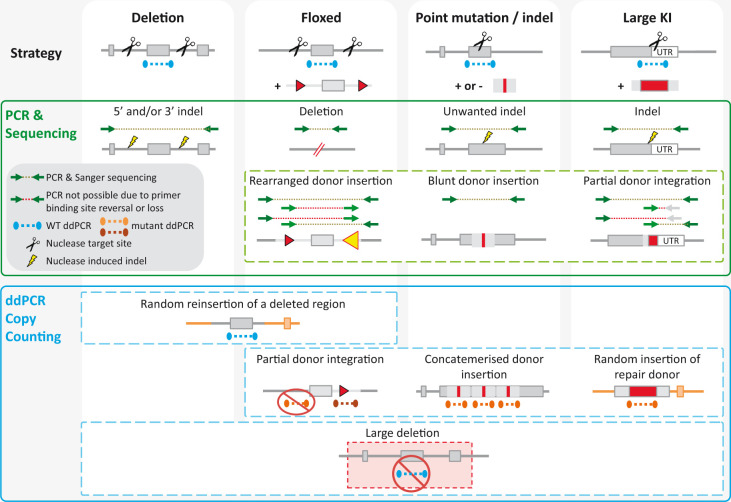
Unintended mutations and methods to detect them. Different types of unintended mutation can occur depending on the editing strategy employed – the number of nuclease target cut sites and whether a repair template is included – while other mutations, such as large deletions, can occur in all cases. Here we present some common examples along with assays which can be used for their detection. Most simply, polymerase chain reaction (PCR) amplification of the targeted locus may identify unwanted insertions or deletion by a shift in band size. Sequencing PCR amplicons can reveal unintended indels and incorrect donor insertions. An inability to amplify an expected product (indicated by a red dotted line) may indicate a rearranged donor insertion (causing incompatible primer orientation) or a partial integration (failure to insert the primer binding region). Copy counting assays, using droplet digital (dd)PCR (or qPCR) are useful for identifying insertion events which are not readily detected by regular PCR. For example, a repair donor or a deleted region can reinsert randomly elsewhere in the genome and ddPCR assays can help to detect this. Concatemerized on target insertion of a donor can be challenging to identify by regular PCR due to amplification bias, while deletions which expand beyond the primers binding sites will be missed entirely. Both are readily detected by ddPCR.