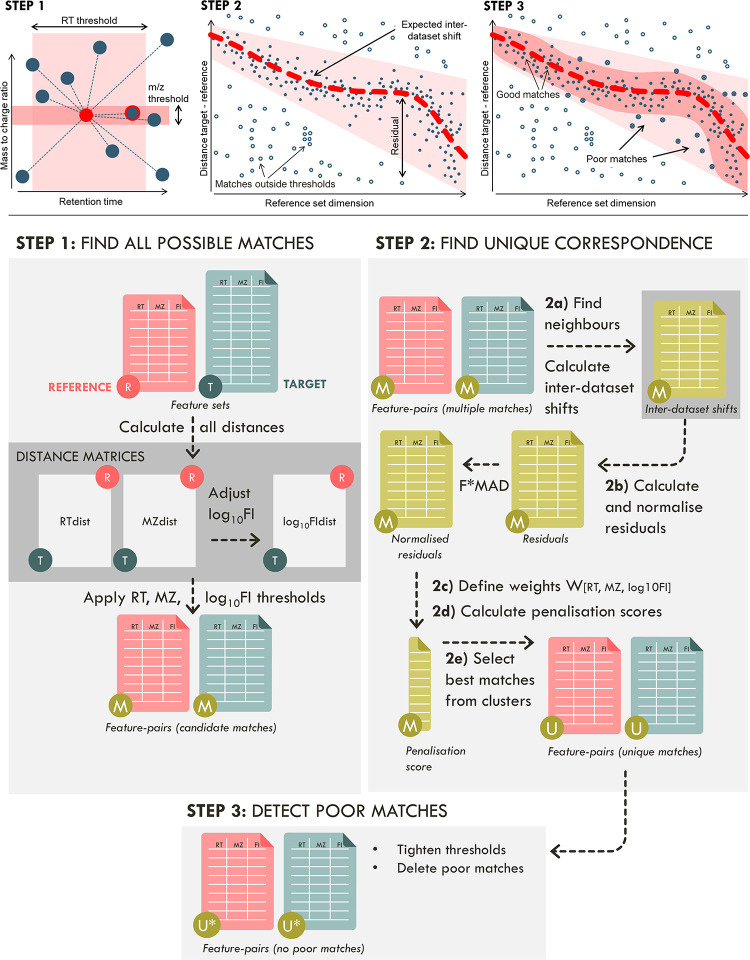
Figure 1.
Method workflow: (top) overview of the approach; (bottom) matrices calculated at each step; Step 1: Distances between all features are calculated (RTdist, MZdist, log10FIdist) and linear thresholds set in all dimensions, finding “M” candidate matches between feature sets; Step 2: Find one-to-one feature correspondence: 2a: The expected inter-dataset shifts are modeled using neighbor consensus; 2b: residuals can then be obtained for each candidate match, normalized; 2c and 2d: transformed into single-value penalization scores; 2e: these are used to define feature-pair matrices containing only “U” unique matches. Step 3. A nonlinear tightening of thresholds is applied to filter out poor matches far from the inter-dataset shifts, yielding “U*” unique matches.