FIGURE 1: Schematic diagram of the steps in a canonical multiplexed tissue imaging experiment and the associated metadata.
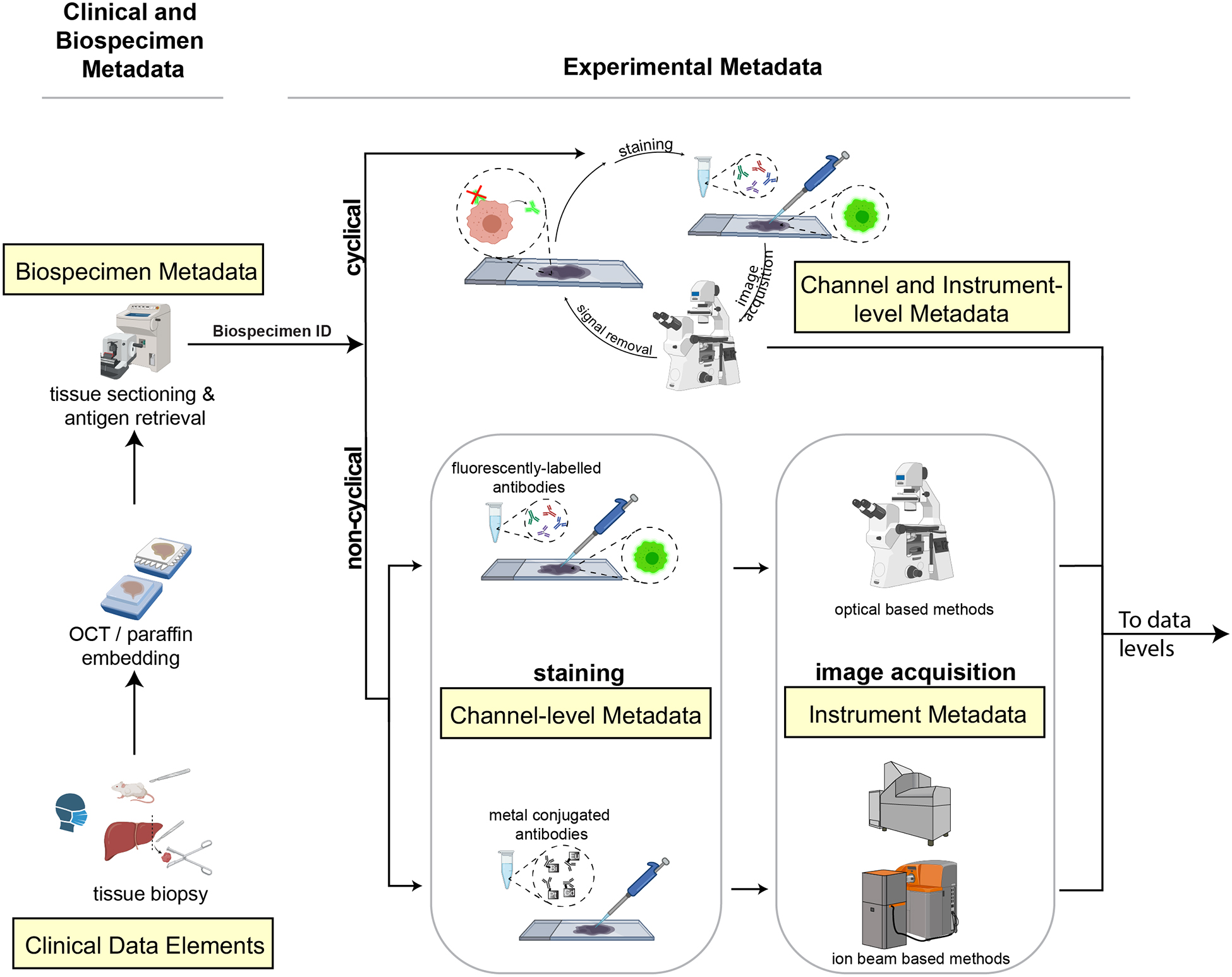
In a typical workflow, samples collected from patient biopsies and resections or from animal models are formaldehyde fixed and paraffin embedded (FFPE) or frozen and then sectioned and mounted onto either a standard glass microscope slide (for CyCIF, mIHC, IMC, MELC or mxIF), fluidic chamber (for CODEX) or specialized carriers (for MIBI). Clinical and biospecimen metadata (extracted from clinical records, for example) is linked to all other levels of metadata via a unique ID (Biospecimen ID). Data is acquired using cyclical or non-cyclical staining and imaging methods and both reagent and experimental metadata collected (consisting of antibody, reagent and instrument metadata). In both cyclic and non-cyclic methods, sections undergo pre-processing, antigen retrieval, and antibody incubation and images are acquired. In cyclical imaging methods, fluorophores or chromogens are inactivated or removed and additional antibodies and/or visualization reagents are applied and data acquisition repeated. Channel and instrument metadata capture these essential details. Created with BioRender.com.
