Extended Data Fig. 1. Cross-population PRS results for real UK Biobank traits, using summary statistics from a meta-analysis of many cohorts.
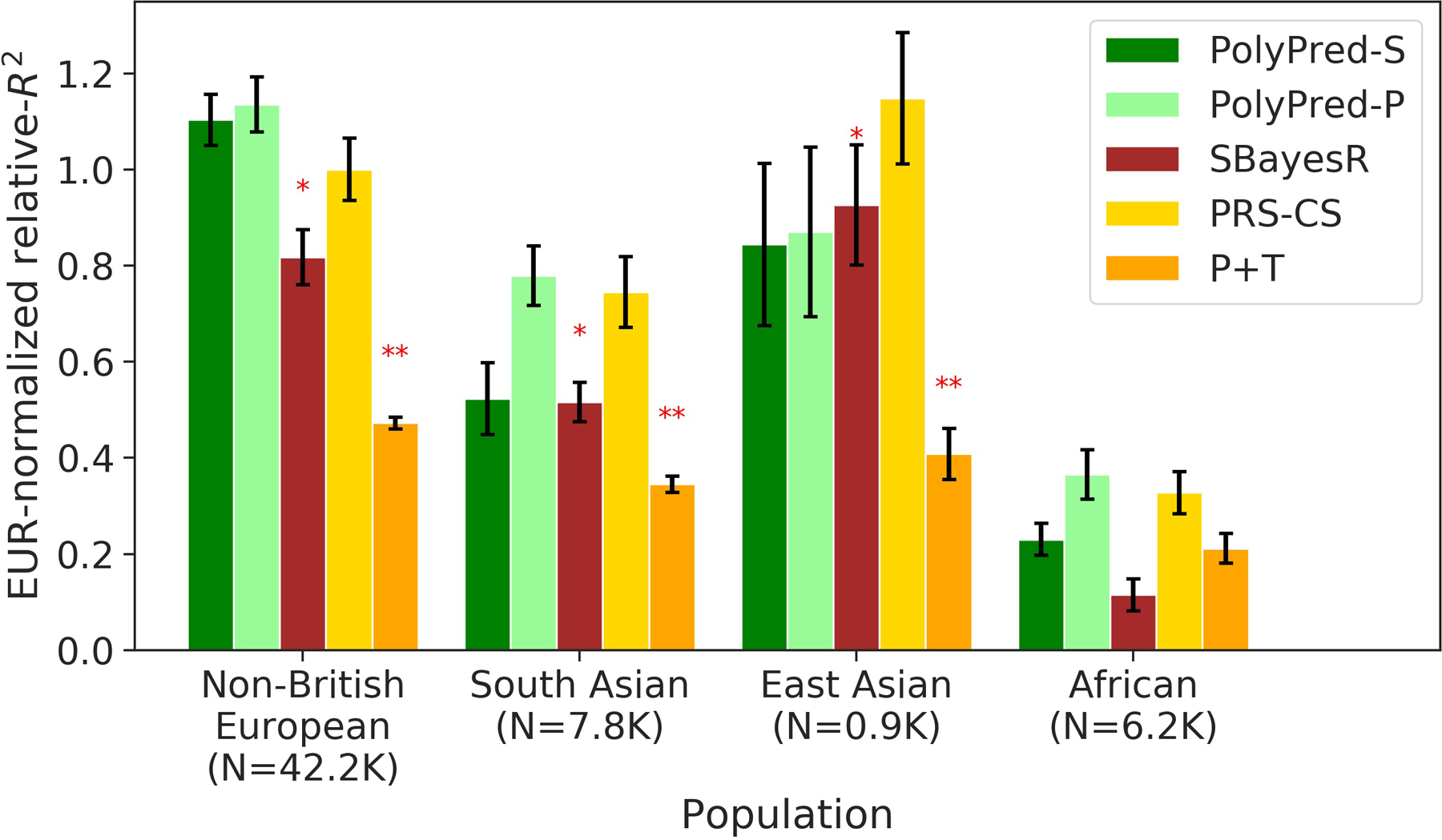
We report average prediction accuracy (relative-R2, but computed with respect to PRS-CS instead of BOLT-LMM; see main text), meta-analyzed across 4 well-powered, approximately independent traits, for PRS trained in European Network for Genetic and Genomic Epidemiology (ENGAGE) samples (average N=61,365) and applied to 4 UK Biobank populations. Target population sample sizes are indicated in parentheses; PolyPred and its summary statistic-based analogues used 500 additional training samples from each target population to estimate mixing weights. Asterisks above each bar denote statistical significance of the difference vs. PRS-CS, with red asterisks denoting a disadvantage (*P<0.05; **P<0.001). P-values were computed using a two-sided Wald test and were not adjusted for multiple comparisons. Errors bars denote standard errors. Numerical results, results for all 4 traits analyzed, absolute prediction accuracies (R2), and P-values of relative improvements vs. PRS-CS are reported in Supplementary Table 5 and Supplementary Table 8.
