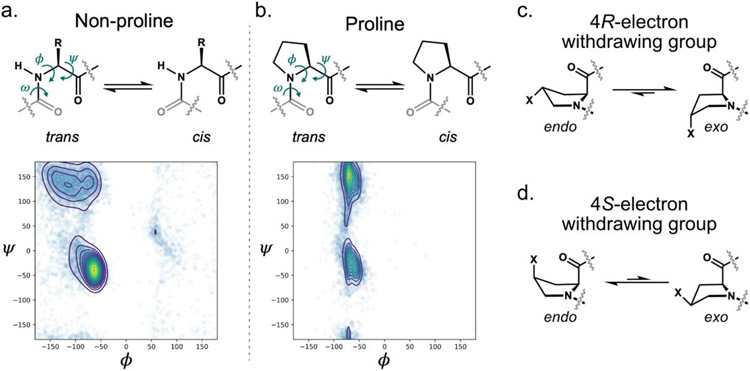
Figure 1.
Proline conformational preferences. a-b. Backbone conformations of and Ramachandran plots for non-proline (a) and proline (b) residues found in structural data deposited in the Protein Data Bank (PDB). The torsional angles ψ, ϕ, and ω, along with cis and trans amide isomers are indicated in the amino acid structures. c-d. Endo and exo ring pucker preferences for proline analogs containing electron withdrawing substituents at the 4 position. ncPro residues with 4R-electron withdrawing groups prefer the exo pucker (c); those with 4S-electron withdrawing groups prefer endo (d).