Table 1:
Proline analogs amenable to residue-specific incorporation into recombinant proteins
| Proline analog |
Proline structure | CAS | ProRS variant |
Reported [NaCl] (M) |
Reported level of proline replacement |
Reference |
|---|---|---|---|---|---|---|
| 4R-OH |
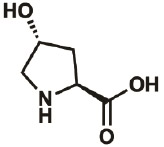
|
51-35-4 | wt | 0.3 | 88% | Lieblich, et al., 2017 |
| 4S-OH |
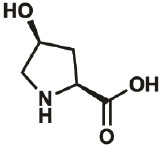
|
618-27-9 | wt | 0.3 | 91% | Lieblich, et al., 2017 |
| 4R-F |
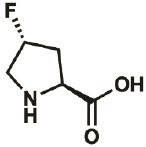
|
2507-61- 1 |
wt | 0.3 | 96% | Lieblich, 2017 |
| 4S-F |
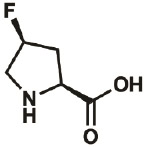
|
2438-57- 5 |
wt | 0.3 | 97% | Lieblich, 2017 |
| 4,4-diF |
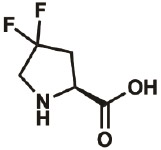
|
52683- 81-5 |
wt | 0.3 | 86% | Lieblich, 2017 |
| 3S-F |
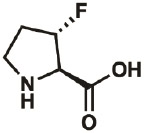 * * |
261350- 70-3 |
– | – | “virtually complete” |
Kim, Hardcastle, & Conticello, 2006 |
| 3R-F |
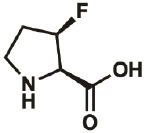
|
261350- 69-0 |
– | – | “virtually complete” |
Kim, Hardcastle, & Conticello, 2006 |
| Dhp |
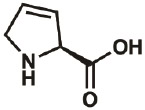
|
4043-88- 3 |
wt | 0.3 | ≤100% | Fang, Lieblich, & Tirrell, 2019 |
| Thz |
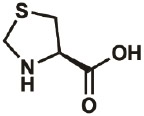
|
34592- 47-7 |
wt | 0.3 | 90% | Fang, Lieblich, & Tirrell, 2019 |
| Aze |

|
2133-34- 8 |
wt | 0.3 | ≥100% | Fang, Lieblich, & Tirrell, 2019 |
| Pip |
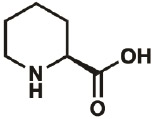
|
3105-95- 1 |
C443G | 0.3 | 89% | Fang, Lieblich, & Tirrell, 2019 |
