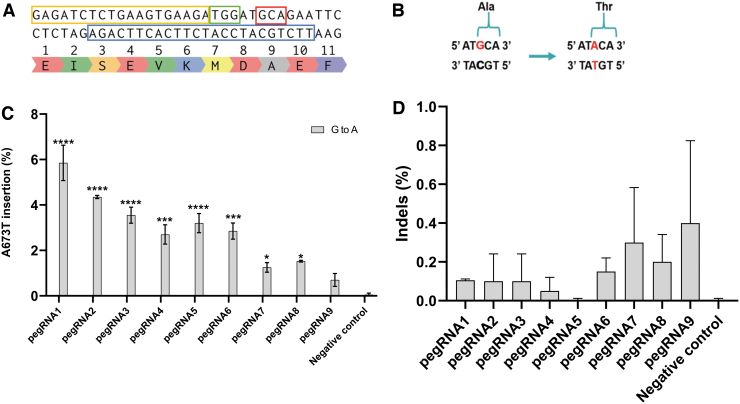
FIG. 1.
Test of various prime editing guide RNAs (pegRNAs) targeting the APP gene in HEK293T cells using the prime editor 2 (PE2) method. (A) Section of APP exon 16 with codon of interest (Ala673) in the red box. In the blue box, the unmodified sequence used to design the primer binding site (PBS) and the reverse transcriptase (RT) template (RTT). The protospacer adjacent motif (PAM) is in the green box, and the pegRNA spacer sequence is in the yellow box. (B) The target nucleotide G (guanine) in red is modified into an A (adenine) in red to change the alanine codon (5′GCA3′) into a threonine codon (5′ACA3′). (C) Nine different pegRNAs with various PBS and RTT were transfected in HEK293T cells with Lipofectamine™ 2000. DNA was extracted 72 h later, and exon 16 of the APP gene was polymerase chain reaction (PCR) amplified. The sequences were then analyzed using the Illumina deep-sequencing method. The negative control did not receive any treatment. The results were analyzed with a one-way analysis of variance (ANOVA) test and Dunnett's multiple comparisons test (n = 2) compared to the negative control. *p < 0.0387; ***p < 0.0002; ****p < 0.0001. (D) Insertion or deletion (indel) levels resulting from prime editing of the APP gene exon 16 using the PE2 method. There were no indels in the control sample. The level of indels varied from 0% to 0.8% for the PE2 method. The results were analyzed with a one-way ANOVA test and Dunnett's multiple comparisons test (n = 2) compared to the negative control. The p-value was not significant for all samples.