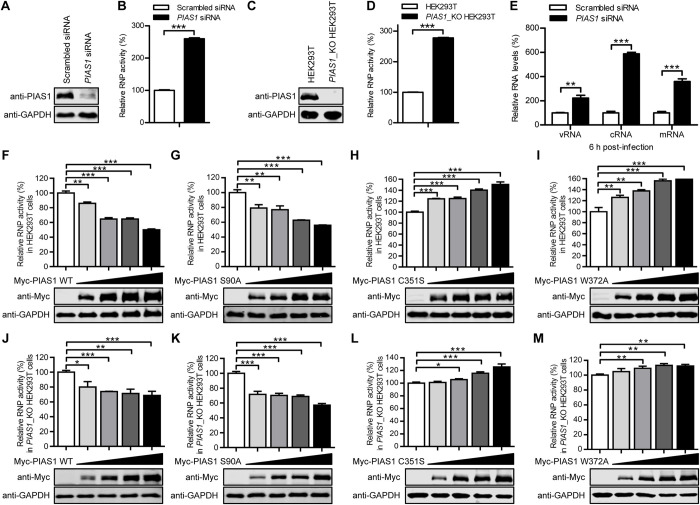
Fig 4. PIAS1 relies on its SUMO E3 ligase activity to suppress IAV transcription and replication.
(A) siRNA knockdown of PIAS1 in HEK293T cells. HEK293T cells were transfected with PIAS1 siRNA or scrambled siRNA for 48 h. Knockdown of PIAS1 expression was confirmed by western blotting with a rabbit anti-PIAS1 pAb. (B) Minigenome assay in siRNA-treated HEK293T cells to determine the effect of endogenous PIAS1 on viral RNP activity. HEK293T cells treated with siRNA as in (A) were transfected with plasmids for the expression of four viral RNP proteins (WSNPB2, WSNPB1, WSNPA, and WSNNP), together with pHH21-SC09NS F-Luc and pRL-TK. Thirty-six hours later, a dual-luciferase assay was performed in which the relative firefly luciferase activity was normalized to the internal control, Renilla luciferase activity. ***, P < 0.001. (C) Establishment of a PIAS1_KO HEK293T cell line. The knockout of PIAS1 was confirmed by western blotting with a rabbit anti-PIAS1 pAb. (D) Minigenome assay in PIAS1_KO HEK293T cells to determine the effect of endogenous PIAS1 on viral RNP activity. The minigenome assay was performed in PIAS1_KO HEK293T cells as in (B). ***, P < 0.001. (E) RT-qPCR analysis to determine the effect of PIAS1 knockdown on the transcription and replication of viral RNAs. A549 cells were transfected with siRNA targeting PIAS1 or with scrambled siRNA for 48 h, followed by infection with WSN (H1N1) virus (MOI = 5). Total RNA was extracted at 6 h p.i. by using TRIzol reagent, and the levels of NP-specific vRNA, cRNA, and mRNA were analyzed by RT-qPCR and then normalized to GAPDH mRNA. The values shown are standardized to the corresponding RNA expression level in the scrambled siRNA-treated A549 cells (100%). **, P < 0.01, ***, P < 0.001. (F-I) Minigenome assay in HEK293T cells to examine the effect of exogenously expressed wild-type PIAS1 and PIAS1 mutants on viral RNP activity. HEK293T cells were transfected with plasmids for the expression of four viral RNP proteins and increasing amounts of Myc-PIAS1 or Myc-PIAS1 mutant, together with pHH21-SC09NS F-Luc and pRL-TK. At 36 h post-transfection, a dual-luciferase assay was performed in which the relative firefly luciferase activity was normalized to the Renilla luciferase activity. **, P < 0.01, ***, P < 0.001. (J-M) Minigenome assay in PIAS1_KO HEK293T cells to examine the effect of exogenously expressed wild-type PIAS1 and PIAS1 mutants on viral RNP activity. The minigenome assay was performed in PIAS1_KO HEK293T cells as in (F-I). *, P < 0.05, **, P < 0.01, ***, P < 0.001.