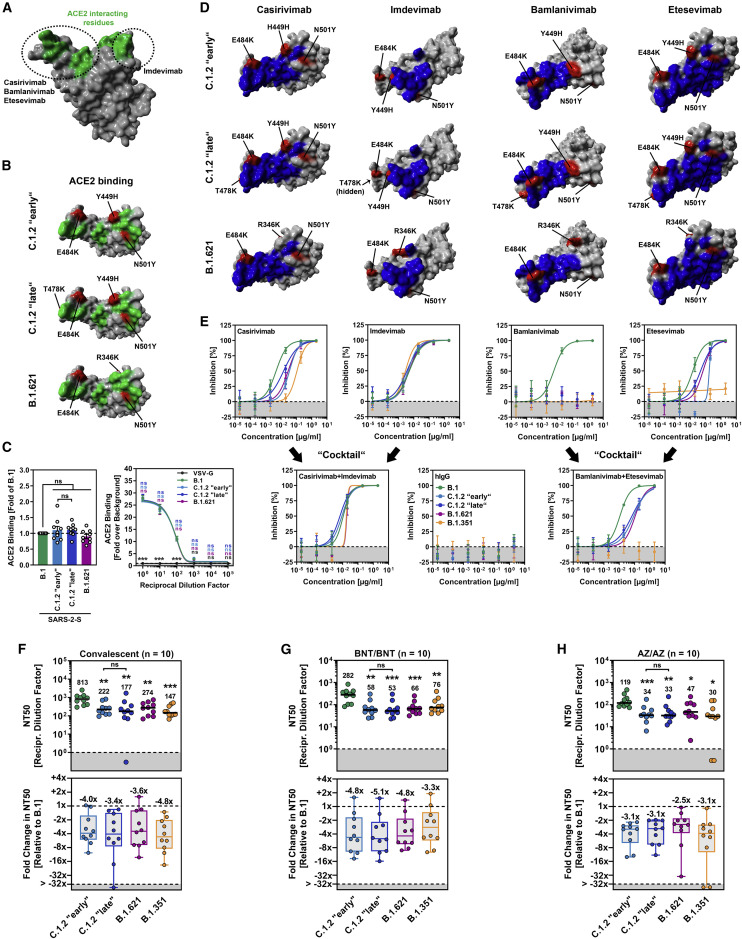
Figure 2.
Spike proteins of SARS-CoV-2 variants C.1.2 and B.1.621 robustly bind ACE2 and efficiently evade antibody-mediated neutralization
(A) Protein model of the SARS-CoV-2 RBD in which the ACE2-interacting interface (green) and target regions of therapeutic monoclonal antibodies (circles) are highlighted.
(B) Location of C.1.2 “early,” C.1.2 “late,” and B.1.621-specific mutations (red) in the context of the RBD (gray). RBD residues that interact with ACE2 are colored in green.
(C) 293T cells expressing the indicated S proteins (or VSV-G) were incubated with soluble ACE2-Fc and secondary antibody and analyzed by flow cytometry (left) and CABA (right). Shown are the average (mean) data from nine (flow cytometry; single samples) or three (CABA, four technical replicates) biological replicates for which ACE2 binding was normalized against B.1 (flow cytometry; set as 1) or the background (CABA; no soluble ACE2-Fc, set as 1). Error bars indicate the SEM. Statistical significance was analyzed by two-tailed Student’s t test with Welch’s correction (flow cytometry) or two-way analysis of variance with Dunnett’s post-hoc test (CABA) (p > 0.05, ns; p < 0.05, ∗). See also Figure S1.
(D) Location of C.1.2 “early,” C.1.2 “late,” and B.1.621-specific mutations (red) in the context of the RBD (gray) epitopes targeted by casirivimab, imdevimab, bamlanivimab, and etesevimab (blue).
(E) Pseudotyped particles bearing the indicated S protein were pre-incubated in with the indicated antibodies or antibody cocktails and subsequently inoculated onto Vero cells. At 16–18 h post inoculation, pseudotype entry was quantified and normalized against samples that did not contain antibody (= 0% inhibition). Shown are the average (mean) data from a single biological replicate (performed with technical quadruplicates), and data were confirmed in a separate independent experiment. Error bars indicate the standard deviation. See also Figure S1.
(F–H) Pseudotyped particles bearing the indicated S proteins were pre-incubated in the presence of convalescent plasma (F) or serum from individuals either vaccinated with BioNTech/Pfizer’s BNT162b2 vaccine (BNT/BNT (G) or AstraZeneca’s ChAdOx1 nCoV-19 vaccine (AZ/AZ) (H), and subsequently inoculated onto Vero cells. At 16–18 h post inoculation, pseudotype entry was quantified and used for the calculation of the neutralizing titer 50 (NT50). Presented are the data from ten plasma/serum samples per group (black lines and numerical values indicate the median NT50). Further, the median fold reduction in NT50 between SARS-CoV-2 B.1 (set as 1) and the indicated variants was calculated (boxplots indicate the median, quartiles, and range; circles indicate individual samples). Statistical significance was analyzed by two-tailed Mann-Whitney test with 95% confidence level (p > 0.05, ns; p ≤ 0.05, ∗; p ≤ 0.01, ∗∗; p ≤ 0.001, ∗∗∗).
See also Figure S2.