Figure 1.
Cryo-EM structure of the SARS-CoV-2 Omicron spike in complex with ACE2
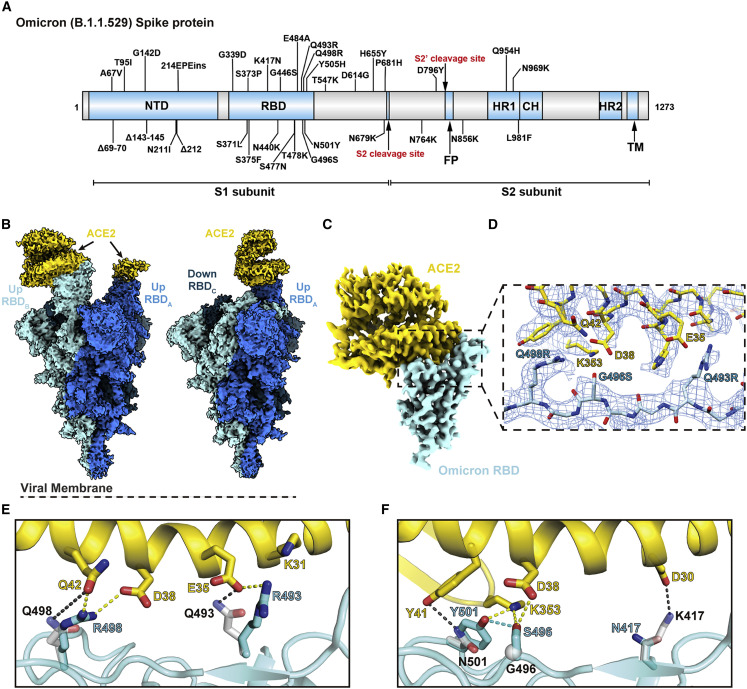
(A) An overview diagram of the domain arrangement of the Omicron spike protein.
(B) Cryo-EM structures of Omicron spike with double- (left) and single (right)-bound ACE2, with three protomers shown in different colors.
(C) Cryo-EM map of the Omicron spike RBD in complex with human ACE2 after local refinement.
(D) Cryo-EM density at the Omicron RBD-ACE2 interface. The fitted atomic model is shown as sticks with oxygen colored in red, nitrogen colored in blue, and sulfurs colored in yellow.
(E and F) Comparison of the RBD-ACE2 interface between WT and the Omicron variant. Residues forming hydrogen bonds and ionic interactions are shown as sticks. Yellow and black dashed lines represent interactions between ACE2 and Omicron or WT RBDs, respectively. The hydrogen bond formed between Omicron RBD mutations G496S and N501Y is shown as a blue dashed line.
See also Figures S1 and S2; Table S1.