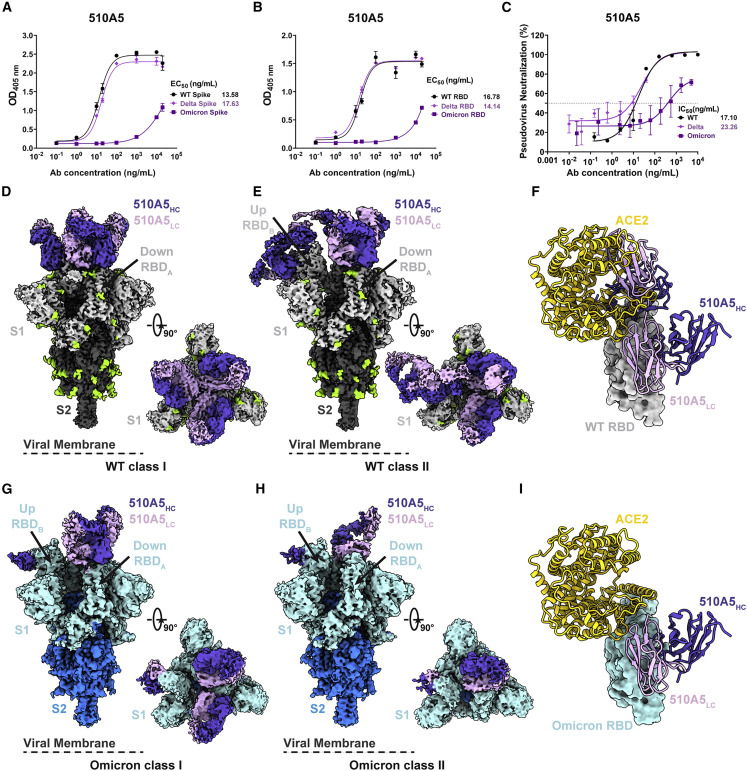
Figure 3.
Cryo-EM structures of 510A5 mAbs binding to SARS-CoV-2 WT, Delta, and Omicron spike protein
(A and B) The binding capabilities of 510A5 against spike protein (A) and RBD protein (B) of WT, Delta, or Omicron measured by ELISA. Data are presented as mean values ± SEM of three independent experiments.
(C) The neutralizing potencies of the 510A5 against WT, Delta, and Omicron measured by pseudovirus neutralization assay. Data are presented as mean values ± SEM of three independent experiments. Dashed line indicates a 50% reduction in viral neutralization.
(D and E) Cryo-EM densities for the 510A5 Fab-WT spike complex observed in two classes. (D) WT class I, 3.2 Å, revealing binding of 510A5 to RBDs in the “3 down” state; (E) WT class II, 3.4 Å, revealing binding of 510A5 to RBDs in “1 up, 2 down” state.
(F) Superposition of the local refined RBD-ACE2 model to that of WT RBD-510A5 model shows binding of two Fabs on both epitopes 1 and 3 completely blocks the ACE2 binding.
(G and H) Cryo-EM densities for the 510A5 Fab-Omicron spike complex observed in two classes with RBDs in a “1 up, 2 down” state. (G) Omicron class I, 3.7 Å, three Fabs bound; (H) Omicron class II, 3.7 Å, two Fabs bound.
(I) Superposition of the local refined Omicron RBD-ACE2 model to that of the 510A5-Omicron RBD model shows no steric hindrance between 510A5 Fab and ACE2.
See also Figures S3 and S4; Table S2.