Figure 4.
Structural mechanism of Omicron spike escaping 510A5
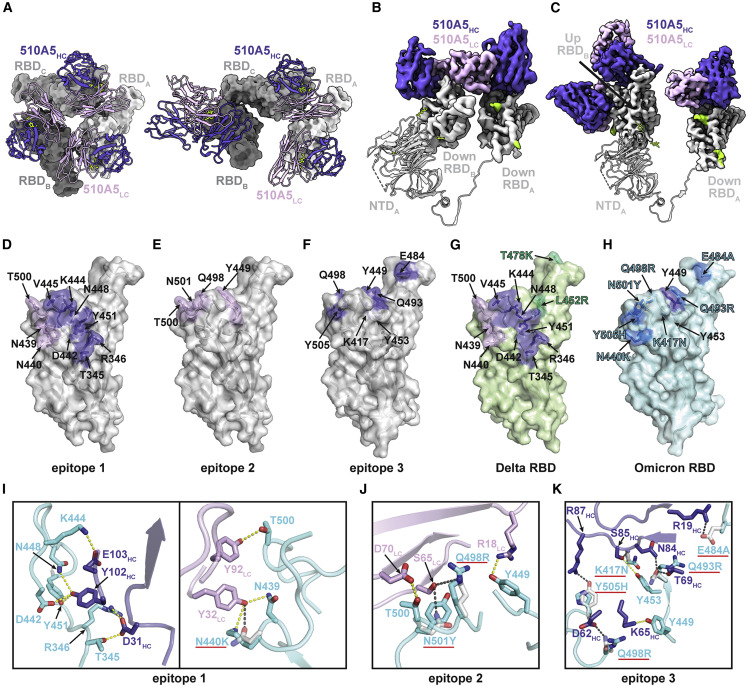
(A) Left, a close-up view of 510A5 light-chain-mediated contacts on adjacent protomer “down” RBD, preserving a 3-fold symmetric conformation in WT class I. Right, a close-up view of an asymmetric conformation with four Fabs bound to the “1 up, 2 down” RBD conformation in WT class II.
(B) 510A5 shares inter-protomer contacts via binding to an adjacent down RBD to form the effective epitope 1 in all five classes of complexes observed.
(C) An extra 510A5 Fab binds to the up RBD to form epitope 3 in WT class II.
(D–F) Surface representations of 510A5 epitope 1 (purple and pink, D), epitope 2 (pink, E) and epitope 3 (purple, F) on the WT RBD surface (gray). RBD epitope residues (defined as residues forming potential hydrogen bonds with 510A5 Fab residues) are labeled in black.
(G) Surface representations of 510A5 epitope 1 (purple and pink) on the Delta RBD surface (green). RBD epitope residues are labeled in black. Two Delta mutations are labeled in green.
(H) Surface representations of the Omicron RBD (blue). Mutations on the Omicron RBD surface region corresponding to the WT epitope 3 are labeled in blue.
(I and J) Composite model of 510A5 Fab-WT RBD overlaid with the 510A5 Fab-Omicron RBD model. Potential hydrogen bonds on epitope 1 (I) and epitope 2 (J) are illustrated by dashed yellow (Omicron) or black (WT) lines, respectively. Omicron mutations are highlighted with red underlines.
(K) Composite model of 510A5 Fab-WT RBD overlaid with the Omicron RBD model. Potential hydrogen bonds are illustrated by dashed yellow (Omicron) or black (WT) lines, respectively. Omicron mutations (highlighted with red underlines) disrupt numerous interactions to 510A5 and exclude the epitope 3.
See also Figure S5.