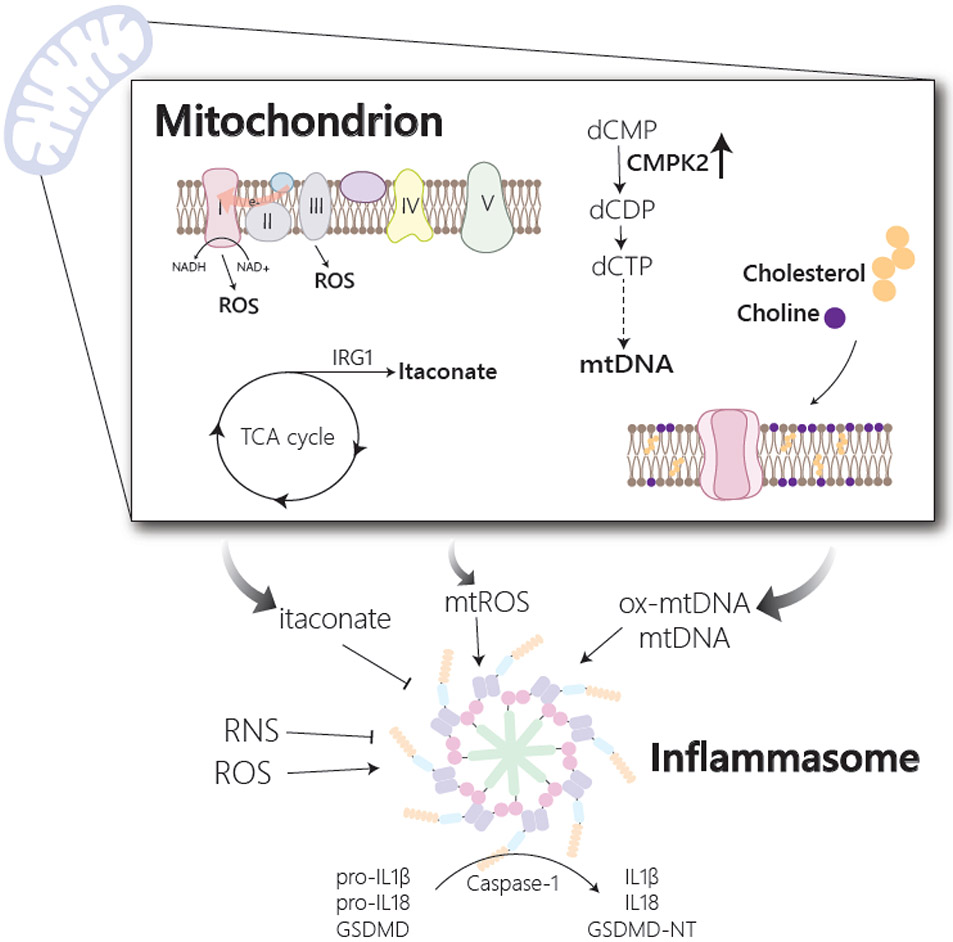
Figure 2. Macrophage metabolism influences inflammasome activation.
Activation of the inflammasome allows for processing of the inflammatory cytokines, IL1β and IL18, and gasdermin D (GSDMD) to their mature and active forms. Inflammasome activation is influenced by changes in multiple metabolic pathways, many of which impinge on mitochondria. Release of mitochondrial DNA (mtDNA) or its oxidized form (ox-mtDNA) from mitochondria serves as an inflammasome activating signal and is regulated by multiple metabolic changes. In response to stimulation, upregulation of the mitochondrial metabolic enzyme, CMPK2, which participates in the synthesis of mitochondrial dCTP and dUTP, supports the production of mtDNA, while alteration in cholesterol and choline production influence mitochondrial membrane composition and mtDNA release. Reactive oxygen species (ROS) from both mitochondrial and cytosolic sources has also been implicated in the activation of the inflammasome. Mitochondrial ROS (mtROS) has been shown to accumulate in response to stimulation and is reported to originate either from complex III of the electron transport chain or from complex I via a phenomenon termed reverse electron transport (RET). In response to activation, there is also an accumulation of reactive nitrogen species (RNS) and the TCA cycle metabolite, itaconate, both of which have been shown to inhibit inflammasome activation.