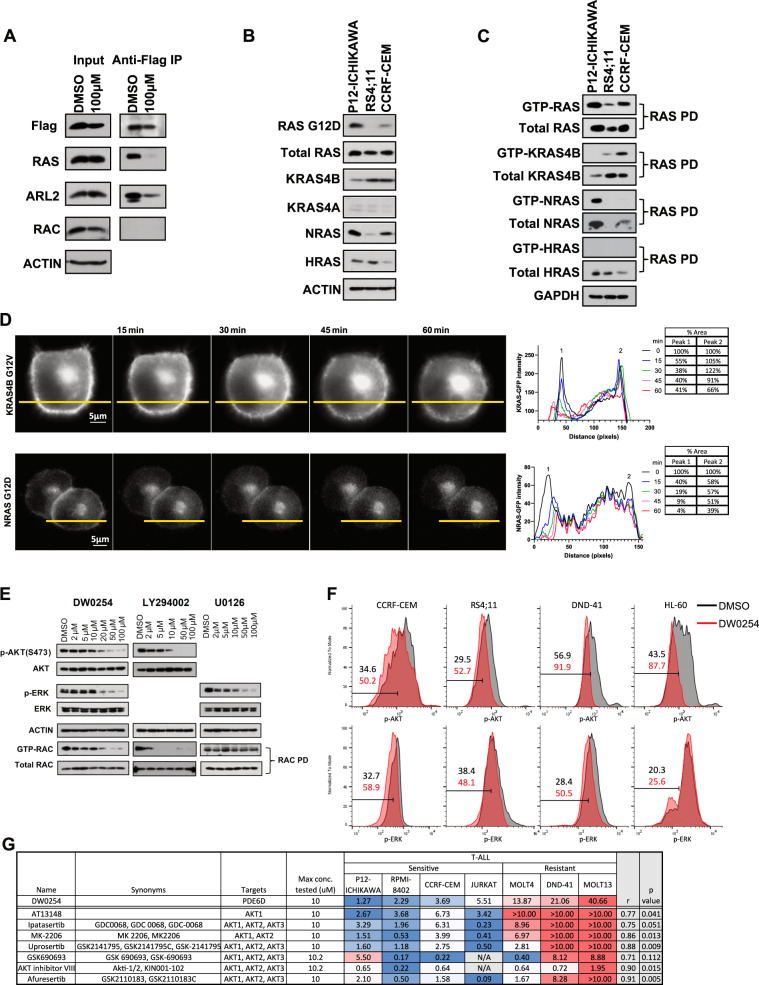
Fig. 6. The expression and activation of RAS isoforms in DW0254 sensitive leukemia cells and the effects of DW0254 on PDE6D/RAS interaction and RAS subcellular location.
A DW0254 treatment inhibits the binding of PDE6D to RAS and ARL2 in P12-ICHIKAWA cells. Co-immunoprecipitation (CoIP) was performed with an anti-FLAG antibody (F1804, Sigma-Aldrich) on lysates from FLAG-tagged PDE6D transduced P12-ICHIKAWA cells treated with 100 µM DW0254 or DMSO. Cell lysates (Input) and protein eluted from beads (IP) were analyzed by Western blotting with anti-Flag, anti-RAS (05-1072, Millipore Sigma, Billerica, MA), anti-ARL2 (ab183510, Abcam, Cambridge, MA), anti-RAC (610651, BD) antibodies. B The expression pattern of RAS isoforms in leukemia cell lines sensitive to DW0254. Western blot analysis of whole-cell lysates from P12-ICHIKAWA, RS4;11, and CCRF-CEM leukemia cells detected by anti-RASG12D (14429S, Cell signaling), anti-RAS (05-1072, Millipore Sigma), Anti-KRAS4B (WH0003845M1, Millipore Sigma), anti-KRAS4A (ABC1442, Millipore Sigma), anti-NRAS (sc-31, Santa Cruz), and anti-HRAS (18295-1-AP, Proteintech, Rosemont, IL) antibodies. C Activated RAS isoforms in P12-ICHIKAWA, CCRF-CEM and RS4;11 cells. GST pulldown assays were performed by incubating protein lysates prepared from P12-ICHIKAWA, CCRF-CEM and RS4;11 with RAF-1 RBD conjugated agarose beads. The GTP-RAS proteins bound to the beads or the whole cell lysates to detect the level of total RAS protein were identified using anti-RAS, anti-KRAS4B, anti-NRAS, or anti-HRAS antibodies described above. For A, B, and C, beta-ACTIN or GAPDH (A300-641A, BETHYL, Montgomery, TX) were used as a protein loading control, one representative experiment of two or three is shown. D Mislocalization from cell surface membrane of GFP-tagged mutant KRAS4BG12V (upper panel) or NRASG12D (lower panel) in PANC-1 cells after treatment with 20 µM of DW0254. Time in minutes is indicated above the panels; the first panel represents the moment immediately after the addition of the inhibitor. The intensity profiles on the right show changes in fluorescence along the X axis, represented by yellow lines on the micrographs. Tables show changes in percentage of the area under the peaks, where time 0 peaks represent the intersection of the X axis with the cell membrane. E Western blot showing total and phosphorylated AKT and ERK, and pulldown results for RAC activation in P12-ICHIKAWA cells treated with increasing doses of DW0254, PI3K inhibitor LY294002, or MEK inhibitor U0126 for 3 h. Total AKT, and ERK expression were assessed using anti-AKT (9272, Cell signaling) and anti-ERK (9102S, Cell signaling) antibody respectively. Phosphorylation of AKT and ERK were assessed using anti-phospho-AKT Ser473 (9271S, Cell signaling) and anti-phospho-ERK (4377S, Cell signaling) antibody respectively. Total RAC and GTP-RAC were analyzed by RAC pull-down assay as described in Fig. 2B. For A, B, C, and D, beta-ACTIN or GAPDH were used as a protein loading control. Data represent three independent experiments. F PhosphoFlow results showing intensity of p-AKT (top) and p-ERK (bottom) intracellular staining on cells treated with DMSO only (black) or 50 µM DW0254 (red) for 1 h. Data are representative of two independent experiments with four cell lines showing the same results. G Summary of response to AKT inhibitors in T-ALL cell lines from “The Genomics of Drug Sensitivity in Cancer Project” database compared to sensitivity to DW0254. Pearson’s correlation coefficient (r) and the p value for the correlation coefficient between response to DW0254 and each AKT inhibitor are shown on the right extremity.