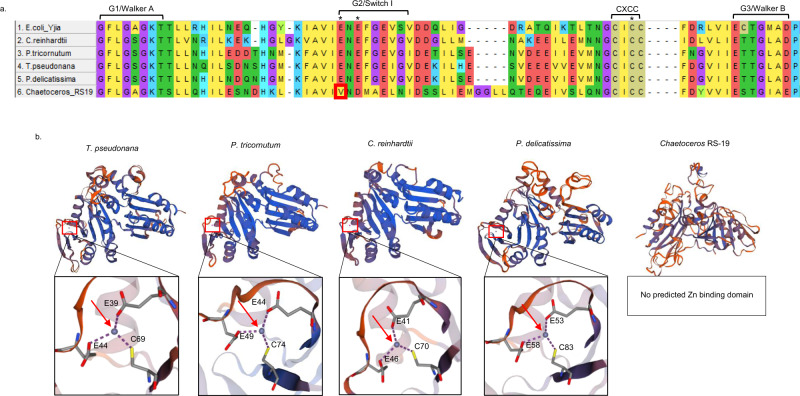
Fig. 3. Algal ZCRP-A sequence alignments and predicted structures.
a Sequence alignment of the E. coli YjiA protein compared to ZCRP-A proteins in C. reinhardtii, P. tricornutum, T. pseudonana, P. delicatissima, and Chaetoceros RS19 generated using MUSCLE. Four conserved GTPase domains including the putative CXCC metal binding motif are labeled. Asterisks denote the glutamate (E) and cysteine (C) residues predicted to bind Zn2+. Notably, one of the metal binding E resides in the G2/Switch I region has been replaced by valine (V; red box) in Chaetoceros RS19. b Predicted structures of ZCRP-A homologs modeled using the well-characterized Zn2+ and Co2+ binding COG0523 GTPase YjiA protein in E. coli as a template (PDB entry 4IXM.2.b). The predicted ligand Zn2+ is indicated by a red arrow. Close-up views show the predicted Zn2+ binding site composed of two glutamate (E) and one cysteine (C) residue.