Figure 1.
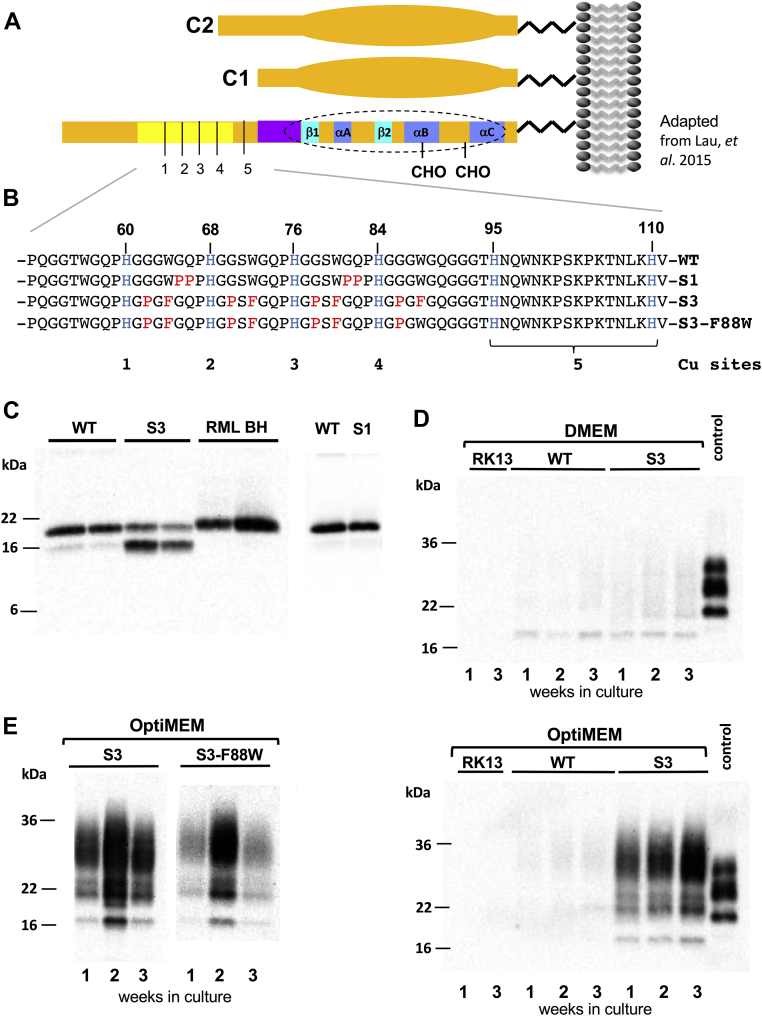
Spontaneous production of protease-resistant PrP.A, schematic of PrP and positions of C1 and C2 fragments. The globular domain is indicated by an extended oval and structures within this (dotted perimeter) include three α-helices and two β-strands, as well as N-glycosylation sites (“CHO”). The OR region is shaded yellow and 1 to 5 refer to Cu-binding sites. B, expanded view of the OR region showing amino acid replacements in the S1, S3, and S3-F88W alleles (modified from Lau et al., 2015 (28)). C, RK13 cells expressing WT-PrP, S3-PrP, or S1-PrP alleles were infected with the RML prion isolate and subjected to PNGase F and PK digestion (50 μg/ml). RML BH refers to biological replicate samples of mouse brain homogenate from infected animals. The origin and identity of the bands in S3 cells (lanes 3 and 4) is elaborated in Figure 3, D and E. D, protease digest comparison of stable clones of WT-PrP and S3-PrP and untransfected RK13 cells grown in two different media, DMEM and Opti-MEM. Cells were grown for 3 consecutive weeks. Cell extracts were treated with 50 μg/ml PK and probed with Sha31 antibody. As a comparison, a brain homogenate of a WT mouse infected with RML is presented (“control”). E, S3 and S3-F88W cells were grown for 3 consecutive weeks in Opti-MEM. Cell lysates were digested with PK as aforementioned. DMEM, Dulbecco's modified Eagle's medium; OR, octarepeat; PK, proteinase K; PNGase F, peptide-N-glycosidase F; PrP, prion protein.