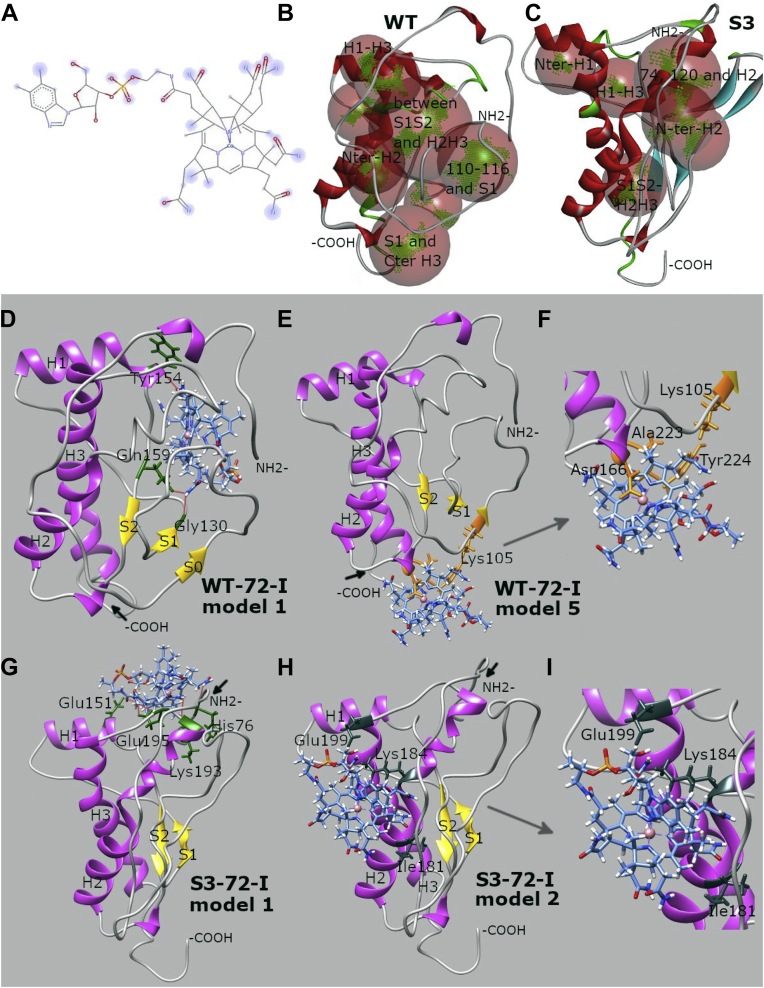
Figure 5.
Docking analysis of PrP–Cbl interactions.A, chemical structure of Cbl with predicted docking sites highlighted in blue. B and C, binding sites in WT and S3 PrP structures (translucent red spheres with areas for compound centering highlighted in green) predicted from receptor cavities by the Biovia Discovery Studio Visualizer. D–I, predicted docking sites of Cbls: WT mPrP 72-I, docking model 1 (D); WT mPrP 72–231-I, docking model 5 (E), and the corresponding close-up view (F); S3 mPrP 72 to 231-I, docking model 1 (G); S3-72-I, docking model 2 (H), and the corresponding close-up view (I). D–I, α-helices of PrP are indicated in purple, β-strands in yellow, and random coils and turns in gray; PrP residues involved in hydrogen bonds are colored green; N and C termini for simplicity are presented as -NH2 and -COOH with the positions of occluded termini indicated by short black arrows; and atom/bonds of Cbl are colored blue with the phosphate group depicted in orange. E and F, in addition illustrate overlapping relationships of Cbl-binding sites with tetrapyrrole binding (globular domain contact residues for tetrapyrrole in orange). H and I, show relationships with Zn-loaded OR region (globular domain contact residues in slate gray) (55, 58), see also Table S5. Cbl, cobalamine; PrP, prion protein; mPrP, mouse prion protein; OR, octarepeat.