Summary
Generated by RNA deprotection or cleavage, 5′ monophosphates trigger RNA degradation in all organisms. Here we describe PABLO-QA (Phosphorylation Assay By Ligation of Oligonucleotides and Quantitative Amplification), a sensitive, low-cost procedure for determining the percentage of specific RNA 5′ ends that are monophosphorylated from their ability to undergo ligation to an oligonucleotide. Comparison to a cognate internal standard and a fully monophosphorylated control allows precise quantification of monophosphorylated 5′ termini by RT-PCR, enabling the analysis of transcripts undetectable by blotting.
For complete details on the use and execution of this protocol, please refer to Richards and Belasco (2021).
Subject areas: Molecular Biology, Gene Expression, Molecular/Chemical Probes
Graphical abstract

Highlights
-
•
PABLO-QA measures the percentage of a given RNA 5′ end that is monophosphorylated
-
•
This electrophoretic assay is reliable, inexpensive, and quantitatively accurate
-
•
The sensitivity of the assay makes it possible to examine even low-abundance RNAs
-
•
This approach enables simultaneous analysis of heterogeneous 5′ termini
Generated by RNA deprotection or cleavage, 5′ monophosphates trigger RNA degradation in all organisms. Here we describe PABLO-QA (Phosphorylation Assay By Ligation of Oligonucleotides and Quantitative Amplification), a sensitive, low-cost procedure for determining the percentage of specific RNA 5′ ends that are monophosphorylated from their ability to undergo ligation to an oligonucleotide. Comparison to a cognate internal standard and a fully monophosphorylated control allows precise quantification of monophosphorylated 5′ termini by RT-PCR, enabling the analysis of transcripts undetectable by blotting.
Before you begin
In both prokaryotic and eukaryotic organisms, rates of RNA processing and degradation are often governed by 5′-terminal deprotection to generate a 5′ monophosphate that triggers subsequent ribonucleolytic attack (Muhlrad et al., 1994; Deana et al., 2008; Richards et al., 2011; Cahová et al., 2015). To enable regulatory pathways of this kind to be dissected, we previously devised a quantitative procedure known as PABLO (Phosphorylation Assay By Ligation of Oligonucleotides) for determining the percentage of a particular RNA 5′ terminus that is monophosphorylated on the basis of the unique ability of such ends to undergo splinted ligation to a synthetic oligonucleotide (Celesnik et al., 2007, 2008; Luciano and Belasco, 2019). A limitation of that method was its reliance on Northern blotting, as many cellular transcripts are present at a concentration that is insufficient for detection on a blot. It also required prior knowledge of the precise location of the 5′ end of interest. We recently modified that assay to improve its sensitivity. Because the new procedure, PABLO-QA (Phosphorylation Assay By Ligation of Oligonucleotides and Quantitative Amplification), involves reverse transcription and PCR amplification, quantification requires spiking in a cognate internal standard whose RT-PCR yield is compared to that of the cellular RNA under investigation (Richards and Belasco, 2021). In addition, to correct for transcript-dependent differences in ligation efficiency, RT-PCR yields are compared before and after treating the RNA sample with an excess of the RNA pyrophosphohydrolase RppH to fully convert each 5′ end to a monophosphate. A benefit of using T4 RNA ligase 1 instead of T4 DNA ligase for PABLO-QA is that it obviates the need for a splint, thereby enabling the simultaneous analysis of heterogeneous RNA 5′ termini, whose exact locations are conveniently determined by sequencing the RT-PCR products.
PABLO-QA also has significant advantages over other methods for measuring the percentage of 5′ ends that are monophosphorylated. Compared to those based on RNA-seq (German et al., 2009; Bischler et al., 2017), it is less costly, generates data that are easier to analyze, and enables accurate measurements even for low-abundance RNAs. In addition, it is quantitatively more reliable than methods based on sensitivity to degradation by 5′-monophosphate-dependent exonucleases such as XRN1 and Terminator (Bandyra et al., 2012), especially when less than half of the 5′ termini under investigation are monophosphorylated.
PABLO-QA involves two stages of analysis. The preliminary stage (i) verifies the efficacy and selectivity of two transcript-specific primers designed by the investigator, (ii) identifies the precise location of the RNA 5′ end(s), and (iii) suggests a suitable location for the 5′ end of a cognate internal standard, which is then synthesized in vitro as a fully monophosphorylated transcript and spiked into the cellular RNA. In the final stage, the percentage of 5′ termini that are monophosphorylated is determined by comparing the band intensities of the RT-PCR products obtained from the RNA 5′ end(s) of interest and the internal standard before and after exhaustive treatment with RppH to convert all of the cellular 5′ ends to monophosphates.
Oligonucleotides
Timing: 1 h
PABLO-QA requires several desalted oligonucleotides, most of which are universal but two of which (primer X and primer Y) are specific for the RNA of interest (see Figure 1 and the key resources table for a diagram and sequences). The RNA-specific primers are designed as follows.
-
1.Design a DNA primer (primer X) for target-specific reverse transcription of the RNA of interest and the first round of PCR.
-
a.This primer needs to anneal far enough downstream of the transcription start site for the product of the first round of PCR to be >100 nt long. This allows for purification of the first-round PCR products on a Qiagen QiaQuick column. A primer length of 18–25 nt, depending on its GC content, is sufficient.
-
b.A possible pitfall in positioning a reverse transcription primer too far from the RNA 5′ end is that it may increase the likelihood of intervening RNA structure that could hinder reverse transcriptase. For that reason, a primer that generates an extension product longer than 250 nt should generally be avoided.
-
a.
-
2.
Design a nested DNA primer (primer Y) for the second round of PCR.
A primer length of 18–25 nt, depending on its GC content, is sufficient. This primer should be designed to anneal 15–50 nt downstream of the 5′ end of the transcript of interest so as to generate one or more PCR products that are sufficiently short (<100 bp) to enable resolution of closely spaced 5′ ends.
Figure 1.
Primers and templates for PABLO-QA
The steps in PABLO-QA after RppH treatment and addition of the internal standard are illustrated. (1) Ligation of the chimeric oligonucleotide (blue and red rectangle) to monophosphorylated RNA (gray line preceded by the letter p). (2) Reverse transcription by extension of primer X (broad white arrow). (3) First round of PCR amplification by extension of primers A (broad blue arrow) and X. (4) Second round of PCR amplification by extension of nested primers B (broad red arrow) and Y (broad black arrow). The products of the second round of PCR amplification are then either examined by gel electrophoresis to compare band intensities and calculate the percentage of 5′ ends that are monophosphorylated (% monoP) or gel purified, individually PCR amplified a third time by extension of primers C (broad green and red arrow) and Y, gel purified again, and sequenced with a primer (primer D, not shown) matching the 5′-terminal segment of primer C to map RNA 5′ ends.
RNA extraction
Timing: 1 day
200–400 μg of total RNA is required to complete the analysis. It can be isolated from bacterial or eukaryotic cells by a number of methods, such as extraction with hot acidic phenol (Luciano et al., 2017), extraction with a phenol/guanidine isothiocyanate reagent (e.g., Invitrogen TRIzol), or elution from a silica matrix (e.g., QIAGEN RNeasy or New England Biolabs Monarch) and must be DNase-treated before use.
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Chemicals, peptides, and recombinant proteins | ||
| RppH | New England Biolabs | M0356S |
| 10× RppH buffer | New England Biolabs | M0356S |
| Phusion high-fidelity DNA polymerase | Thermo Fisher Scientific | F530S |
| 5× Phusion polymerase buffer | Thermo Fisher Scientific | F530S |
| Taq DNA polymerase | New England Biolabs | M0273L |
| 10× Taq polymerase buffer | New England Biolabs | M0273L |
| T7 RNA polymerase | Thermo Fisher Scientific | EP0111 |
| 10× T7 RNA polymerase buffer | Thermo Fisher Scientific | EP0111 |
| T4 RNA ligase 1 | New England Biolabs | M0204S |
| 10× T4 RNA ligase buffer | New England Biolabs | M0204S |
| RNasin | Promega | N251B |
| SuperScript IV reverse transcriptase | Thermo Fisher Scientific | 18080-044 |
| 5× 1st strand buffer | Thermo Fisher Scientific | 18080-044 |
| TURBO DNase | Thermo Fisher Scientific | AM2239 |
| 10× Turbo DNase buffer | Thermo Fisher Scientific | AM2239 |
| Pyrophosphatase, inorganic (yeast) | New England Biolabs | M2403L |
| ATP | Roche | 11140965001 |
| GTP | Roche | 11140957001 |
| CTP | Roche | 11140922001 |
| UTP | Roche | 11140949001 |
| GMP | Millipore Sigma | G8377-5G |
| Deoxynucleotide (dNTP) solution mix | New England Biolabs | N0447L |
| tRNA from E. coli | Millipore Sigma | 10109541001 |
| Poly(A) | Millipore Sigma | P9403 |
| Phenol/chloroform/isoamyl alcohol pH 4.3 | Thermo Fisher Scientific | BP1754I-100 |
| Phenol/chloroform/isoamyl alcohol pH 6.7 | Thermo Fisher Scientific | BP1752I-100 |
| 40% Acrylamide:bis-acrylamide (19:1) | Thermo Fisher Scientific | J60909 |
| Ammonium persulfate | Thermo Fisher Scientific | BP179-100 |
| TEMED | Thermo Fisher Scientific | BP150-20 |
| Urea | Thermo Fisher Scientific | AAJ75826A7 |
| Critical commercial assays | ||
| Qiagen QiaQuick PCR purification kit | QIAGEN | 28106 |
| Qiagen QiaQuick gel purification kit | QIAGEN | 28706 |
| Oligonucleotides | ||
| Chimeric oligonucleotide: CGACTGGAGCACGAGGACACTGACATGGA CTGAAGGAGTAGrArArA |
Integrated DNA Technologies | N/A |
| Primer A:CGACTGGAGCACGAGGACACTGA | Integrated DNA Technologies | N/A |
| Primer B: ACATGGACTGAAGGAGTA |
Integrated DNA Technologies | N/A |
| Primer C: TCATCGTCGCGCTCCAGCGAAAGCGGTCC TCGCCGAAAATGACCCAGAGCGCTG CCGG CAGGACACTGACATGGACTGAAGGAGTA |
Integrated DNA Technologies | N/A |
| Primer D: CATCGTCGCGCTCCAGCG |
Integrated DNA Technologies | N/A |
| Software and algorithms | ||
| Image Lab 6.1 | Bio-Rad | 12012931 |
Materials and equipment
Non-denaturing sample buffer
| Reagent | Final concentration | Amount |
|---|---|---|
| 500 mM EDTA (pH 8.0) | 10 mM | 1 mL |
| Glycerol | 50% | 25 mL |
| Bromophenol blue | 0.1% | 50 mg |
| Water | 24 mL | |
| Total | 50 mL |
Store at room temperature for up to one year.
Formamide sample buffer
| Reagent | Final concentration | Amount |
|---|---|---|
| 500 mM EDTA (pH 8.0) | 20 mM | 2 mL |
| Glycerol | 1% | 500 μL |
| Bromophenol blue | 0.05% | 25 mg |
| Formamide | 95% | 47.5 mL |
| Total | 50 mL |
Store at –20°C for up to one year.
Step-by-step method details
Preliminary analysis
RppH treatment
Timing: 4 h
Exhaustive pretreatment of the cellular RNA with RppH generates a control sample in which the 5′ end of the transcript of interest is fully monophosphorylated. In the preliminary stage of analysis, this allows the visualization of transcripts that are predominantly triphosphorylated or capped. Additionally, in the final, quantitative stage of analysis, this control enables the percentage of RNA 5′ ends that are monophosphorylated to be determined by making it possible to correct mathematically for transcript-dependent differences in ligation efficiency.
-
1.
Assemble the RppH reaction mixtures.
| Reagent | 1 |
2 |
|---|---|---|
| Mock treatment (μL) | RppH treatment (μL) | |
| Total cellular RNA (1 μg/μL) | 7.5 | 7.5 |
| RNasin (40 U/μL) | 0.5 | 0.5 |
| 10× RppH buffer | 2.0 | 2.0 |
| RppH (5 U/μL) | – | 1.2 |
| 1× RppH buffer | 1.2 | – |
| Water | 8.8 | 8.8 |
| Total | 20.0 | 20.0 |
-
2.
Incubate at 37°C for 2 h.
-
3.Add 130 μL of 3 mM EDTA (pH 8.0), extract with phenol/chloroform (pH 4.3), and ethanol precipitate the RNA.
-
a.Add an equal volume of phenol/chloroform (pH 4.3) and mix by shaking.
-
b.Centrifuge at 15,000×g for 5 min at room temperature. Collect the aqueous (upper) layer and transfer it to a new microcentrifuge tube.
-
c.Add 1/10th volume of 3 M sodium acetate (pH 5.2) and 3 volumes of ethanol. Vortex and incubate at −20°C for 15 min.
 Pause point: At this point the samples can be stored indefinitely at −20°C.
Pause point: At this point the samples can be stored indefinitely at −20°C. -
d.Centrifuge at 15,000×g for 30 min at 4°C and discard the supernatant.
-
e.Add 800 μL of 70% ethanol and centrifuge at 15,000×g for 10 min at 4°C.
-
f.Discard the supernatant and air dry the RNA pellets until no liquid is visible.
-
a.
-
4.
Dissolve the RNA pellets in 10 μL of sterile water.
Pause point: At this point the samples can be stored indefinitely at −20°C.
Ligation of a chimeric oligonucleotide
Timing: 4 h
This step uses T4 RNA ligase 1 to ligate a chimeric oligonucleotide to the 5′ end of transcripts that are monophosphorylated (Figure 1). Comprising 41 deoxyribonucleotides and three 3′-terminal ribonucleotides, this chimeric oligonucleotide is long enough to encompass the sequences of two nested forward primers (primers A and B) (key resources table). The three ribonucleotides at its 3′ end are required for T4 RNA ligase 1 to efficiently ligate the oligonucleotide to 5′-monophosphorylated RNA, while the inclusion of deoxyribonucleotides lowers the cost of synthesis.
-
5.
Assemble the ligation mixtures for the mock-treated and RppH-treated cellular RNA samples.
| Nucleic acid mixture | |
|---|---|
| Reagent | Amount for one reaction (μL) |
| Mock- or RppH-treated cellular RNA (0.75 μg/μL) | 10.0 |
| Chimeric oligonucleotide (1 μg/μL) | 1.0 |
| Subtotal | 11.0 |
-
6.
Incubate at 65°C for 5 min and then transfer to ice for 1 min.
-
7.
Add 14 μL of ligase master mix to each tube.
| Ligase master mix | |
|---|---|
| Reagent | Amount for one reaction (μL) |
| Water | 9.25 |
| 10× T4 RNA ligase buffer | 2.5 |
| ATP (100 mM) | 0.25 |
| RNasin (40 U/μL) | 0.5 |
| T4 RNA ligase 1 (10 U/μL) | 1.5 |
| Subtotal | 14.0 |
| Reagent | Amount for one reaction (μL) |
|---|---|
| Nucleic acid mixture | 11.0 |
| Ligase master mix | 14.0 |
| Total | 25.0 |
-
8.
Mix thoroughly and incubate at 37°C for 2 h.
-
9.
Add 125 μL of 3 mM EDTA (pH 8.0), extract with phenol/chloroform (pH 4.3), and ethanol precipitate as described in step 3. Dissolve the RNA pellets in 11.5 μL of sterile water.
Pause point: At this point the samples can be stored indefinitely at −20°C.
Reverse transcription
Timing: 2 h
Reverse transcription with a transcript-specific primer generates cDNA complementary to a 5′-terminal segment of the RNA of interest and the chimeric oligonucleotide to which the RNA has been ligated (Figure 1).
| Nucleic acid mixture | |
|---|---|
| Reagent | Amount for one reaction (μL) |
| Ligated RNA | 11.5 |
| Primer X (10 pmol/μL) | 0.5 |
| dNTP mix (10 mM each) | 1.0 |
| Subtotal | 13.0 |
-
10.
Transfer the ligated RNA sample to a 0.2-mL thin-walled PCR tube and mix with primer X and the dNTP mix. Incubate at 65°C for 5 min and transfer to ice for 1 min.
-
11.
Prepare a master mix containing reverse transcriptase buffer, DTT, RNasin, and reverse transcriptase, and combine it with the nucleic acid mixture.
| Reverse transcriptase master mix | |
|---|---|
| Reagent | Amount for one reaction (μL) |
| 5× 1st strand buffer | 4.0 |
| DTT (0.1 M) | 1.0 |
| RNasin (40 U/μL) | 0.5 |
| Superscript IV (200 U/μL) | 1.5 |
| Subtotal | 7.0 |
| Reagent | Amount for one reaction (μL) |
|---|---|
| Nucleic acid mixture | 13.0 |
| Reverse transcriptase master mix | 7.0 |
| Total | 20.0 |
-
12.
Transfer the tube to a thermocycler with a heated lid. Incubate at 55°C for 1 h and then at 70°C for 15 min.
Pause point: At this point the samples can be stored indefinitely at −20°C.
Amplification
Timing: 6 h
Two rounds of PCR with nested primers (Figure 1) allows the detection of low-abundance cellular RNAs while achieving a high level of transcript specificity.
-
13.Perform the first round of PCR.
-
a.Assemble the PCR mixtures.
Reagent Amount for one reaction (μL) Primer A (10 pmol/μL) 2.5 Primer X (10 pmol/μL) 2.5 10× Taq polymerase buffer 5.0 dNTP mix (10 mM each) 1.0 cDNA sample 1.5 Water 36.5 Taq DNA polymerase (5 U/μL) 1.0 Total 50 -
b.Use a thermocycler with a heated lid and programmed as follows.
Temperature Time Cycles 95°C 1 min 1 95°C 30 s 30 58°C 30 s 72°C 30 s 72°C 5 min 1
-
a.
-
14.
Remove the primers for the first round by purifying the PCR reaction products with a Qiagen QiaQuick PCR purification kit according to the manufacturer’s instructions and eluting the DNA from the column with sterile water.
-
15.Perform the second round of PCR.
-
a.Assemble the PCR mixtures.
Reagent Amount for one reaction (μL) Primer B (10 pmol/μL) 2.5 Primer Y (10 pmol/μL) 2.5 10× Taq polymerase buffer 5.0 dNTP mix (10 mM each) 1.0 Purified 1st round PCR product 5.0 Water 33.0 Taq DNA polymerase (5 U/μL) 1.0 Total 50 -
b.Use the same thermal cycling conditions as for the first round PCR, as described in step 13.
 Pause point: Samples can be stored indefinitely at −20°C after either of the PCR steps.
Pause point: Samples can be stored indefinitely at −20°C after either of the PCR steps.
-
a.
Electrophoresis
Timing: 5 h
Electrophoresis on a non-denaturing 12% polyacrylamide gel allows the PCR products to be separated even if derived from closely spaced transcription start sites or RNA cleavage sites (Figure 1).
-
16.
Cast a non-denaturing 12% polyacrylamide gel.
Note: We routinely use a V16 vertical gel system with a 19-well comb and a gel thickness of 1.5 mm.
| Reagent | Amount |
|---|---|
| 40% Acrylamide:Bis-Acrylamide (19:1) | 15 mL |
| 10× TBE | 5 mL |
| Water | 30 mL |
| 10% Ammonium persulfate | 500 μL |
| TEMED | 40 μL |
-
17.
Use 1× TBE as the gel running buffer and pre-run the polyacrylamide gel for 30 min at 100 V.
-
18.
Add 5 μL of non-denaturing sample buffer containing bromophenol blue to each PCR reaction product.
-
19.
Load 40 μL of each sample onto the gel. Run at 100 V for 15 min or until the dye has entered the gel, and then increase the voltage to 150 V.
-
20.
Stop electrophoresis once the bromophenol blue has reached the bottom of the gel.
-
21.
Stain the gel with 0.5 μg/mL ethidium bromide in 1× TBE for 30 min with gentle agitation.
-
22.
Visualize and photograph the PCR products with a UV transilluminator and excise the bands of interest for sequencing.
5′ end mapping
Timing: 2 days
PCR is used to amplify the small amount of DNA extracted from the polyacrylamide gel and to lengthen the segment upstream of the junction between the ligated oligonucleotide and the RNA (Figure 1). This allows sequencing of the RNA 5′ end that was ligated to the chimeric oligonucleotide, which otherwise would be too close to the end of the PCR product.
-
23.Extract DNA from the excised polyacrylamide gel slice(s).
-
a.Place the excised gel slice into a 1.5-mL microcentrifuge tube and crush the slice with a 1-mL pipette tip. Rolling the tip around the wall of the microcentrifuge tube breaks the polyacrylamide into small pieces.
-
b.Add 300 μL or more of 1 mM EDTA (pH 8.0) to the tube, enough to fully submerge the gel pieces. Vortex and incubate for 16 h at 4°C.
-
c.Pellet the gel fragments by centrifugation at 15,000×g at 4°C for 30 min. Use a pipette to carefully transfer the supernatant to another microcentrifuge tube. If the supernatant still contains small gel pieces, repeat the centrifugation and transfer.
-
d.Add an equal volume of phenol/chloroform (pH 7.6) to the supernatant and mix by shaking.
-
e.Centrifuge at 15,000×g for 5 min at room temperature. Transfer the aqueous layer to another microcentrifuge tube.
-
f.Add 1/10th volume of 3 M sodium acetate (pH 5.2) and 3 volumes of ethanol. Vortex briefly and incubate at −20°C for 15 min.
 Pause point: At this point the samples can be stored indefinitely at −20°C.
Pause point: At this point the samples can be stored indefinitely at −20°C. -
g.Centrifuge at 15,000×g for 30 min at 4°C and discard the supernatant.
-
h.Add 800 μL of 70% ethanol and centrifuge at 15,000×g for 10 min at 4°C.
-
i.Discard the supernatant and air dry the precipitate until no liquid is visible.
-
j.Dissolve the DNA precipitate in 10 μL of water.
-
a.
-
24.Amplify and extend each purified PCR product by performing another round of PCR with universal primer C (key resources table) and transcript-specific primer Y.
-
a.Assemble the PCR mixtures.
Reagent Amount for one reaction (μL) Primer C (10 pmol/μL) 2.5 Primer Y (10 pmol/μL) 2.5 10× Taq polymerase buffer 5.0 dNTP mix (10 mM each) 1.0 Extracted PCR product 2.0 Water 36.0 Taq DNA polymerase (5 U/μL) 1.0 Total 50 -
b.Use the same thermal cycling conditions as for the first round PCR, as described in step 13.
-
a.
-
25.
Resolve the PCR products on a horizontal 1.8% agarose gel in 1× TBE alongside an appropriate DNA size ladder. Stain the gel with ethidium bromide, visualize the PCR products with a UV transilluminator, and excise the bands of interest.
-
26.
Extract the PCR products from the gel slices by using a Qiagen QiaQuick gel purification kit according to the manufacturer’s instructions.
-
27.
Using primer D (key resources table), sequence the PCR products to identify the 5′ end of each of the original RNAs.
Preparation of an internal standard
Timing: 2 days
Determining the percentage of the 5′ ends of interest that are monophosphorylated requires quantitative comparison to a monophosphorylated internal standard that can be reverse transcribed and amplified with the same set of primers. The RT-PCR product of this cognate internal standard must be well resolved from the other RT-PCR products. Visualization of the products of the preliminary round of PABLO-QA on a polyacrylamide gel makes it possible to identify a clear zone on the gel above the RT-PCR products arising from cellular RNA. An internal standard whose 5′ terminus is located 10–50 nucleotides upstream of the 5′ end of the longest cellular transcript under investigation should generate an RT-PCR product that migrates there.
-
28.Use PCR to generate a DNA template for synthesizing the internal standard by in vitro transcription (Figure 2).
-
a.The internal standard should be cognate to the cellular transcript under investigation, but with a 5′ extension.
-
b.The template for PCR can be a cell suspension or a cloned DNA fragment encoding the RNA of interest.
-
c.The forward PCR primer (primer E) should incorporate a T7 promoter (TAATACGACTCACTATAG, underlined) upstream of the intended transcription start site (boldface G). This promoter should be preceded by 5 nucleotides and followed by about 20 transcribed nucleotides complementary to the PCR template.
-
d.Use primer X as the reverse PCR primer.
-
e.Assemble the PCR mixture.
Reagent Amount (μL) Primer E (10 pmol/μL) 2.5 Primer X (10 pmol/μL) 2.5 5× Phusion polymerase buffer 10.0 dNTP mix (10 mM each) 1.0 DNA template 0.5 Water 33.0 Phusion DNA polymerase (2 U/μL) 0.5 Total 50 -
f.Use a thermocycler with a heated lid and programmed as follows.
Temperature Time Cycles 98°C 1 min 1 98°C 30 s 30 55°C 30 s 72°C 30 s 72°C 5 min 1 -
g.Resolve the PCR products by electrophoresis on a 1.2%–1.8% agarose gel in 1× TBE alongside an appropriate DNA size ladder. Stain the gel with ethidium bromide, visualize the PCR products with a UV transilluminator, and excise the band of interest. Extract the PCR product from the gel slice by using a Qiagen QiaQuick gel purification kit according to the manufacturer’s instructions.
-
h.For use as a template for in vitro transcription, the concentration of the purified PCR product should be >20 ng/μL. If necessary, its concentration can be increased by evaporation (e.g., on a SpeedVac concentrator).
-
a.
-
29.Synthesize the monophosphorylated internal standard by in vitro transcription with T7 polymerase in the presence of a 30-fold molar excess of GMP over GTP (Figure 2).
-
a.Assemble the reaction mixture.
Reagent Final concentration Amount 10× T7 RNA polymerase buffer 1× 10.0 μL DNA template (20–100 ng/μL) 10–50 ng/μL 50.0 μL GMP (75 mM) 7.5 mM 10.0 μL GTP (50 mM) 0.25 mM 0.5 μL ATP (100 mM) 1 mM 1.0 μL UTP (100 mM) 1 mM 1.0 μL CTP (100 mM) 1 mM 1.0 μL RNasin (40 U/μL) 0.4 U/μL 1.0 μL Water 13.5 μL Inorganic pyrophosphatase (0.1 U/μL) 2 U/mL 2.0 μL T7 RNA polymerase (50 U/μL) 1,000 units/mL 10.0 μL Total 100 μL -
b.Incubate the reaction mixture at 37°C for 4–12 h.
-
c.Degrade the DNA template by adding 11 μL of 10× Turbo DNase buffer and 4 μL (8 U) of Turbo DNase to the RNA synthesis reaction. Mix well and incubate at 37°C for 1–2 h.
-
a.
-
30.Purify the in vitro transcript on a 6% denaturing polyacrylamide gel.
-
a.Cast a denaturing 6% polyacrylamide-urea gel.Note: We routinely use a V16 vertical gel system with a 19-well comb and a gel thickness of 1.5 mm.Note: This method of purification involves loading the products of in vitro transcription in two wells of unequal width. If a comb that creates a broad well for preparative electrophoresis is not available, then two or three narrower teeth of a regular comb can be taped together for that purpose.
Reagent Amount Urea 24 g 40% Acrylamide:Bis-Acrylamide (19:1) 7.5 mL 10× TBE 5 mL Water To 50 mL 10% Ammonium persulfate 500 μL TEMED 40 μL -
b.Use 1× TBE as the running buffer for electrophoresis and pre-run the polyacrylamide gel for 30 min at 100 V.
-
c.Add 220 μL of formamide sample buffer to the in vitro transcription reaction, heat at 95°C for 5 min, and transfer to ice.
-
d.Wash the unpolymerized acrylamide and urea out of the wells of the gel and load the denatured RNA sample into two wells: 20 μL in a narrow well (the marker lane) and the remainder (∼315 μL) in a broad neighboring well (the preparative lane).
-
e.Run the gel at 100 V for 15 min or until the dye has entered the gel, and then increase the voltage to 180 V. Stop electrophoresis when the bromophenol blue has reached the bottom of the gel.
-
f.Cut the gel vertically between the two lanes and stain the marker lane with 0.5 μg/mL ethidium bromide in 1× TBE for 30 min with gentle agitation. Cover the remainder of the gel with plastic wrap or a plastic sheet protector to prevent it from drying out.
-
g.Place the stained part of the gel on a sheet of clear plastic such as a sheet protector. Visualize the RNA band with a UV transilluminator and use a pen to mark the upper and lower boundaries of the band on the sheet protector.
-
h.Place the unstained portion of the gel on the marked sheet protector so as to align it with the marker lane. Use the outline of the marker band as a guide for excising the RNA band from the unstained part of the gel. This method allows the RNA product to be gel purified without exposing it to ethidium bromide or UV light.
-
i.Extract the RNA from the polyacrylamide gel slice by the method described in step 23, substituting phenol/chloroform (pH 4.3) for phenol/chloroform (pH 7.6).
-
j.After ethanol precipitating and drying the RNA, dissolve it in 40 μL of sterile water and determine its concentration by measuring the absorbance at 260 nm.
-
a.
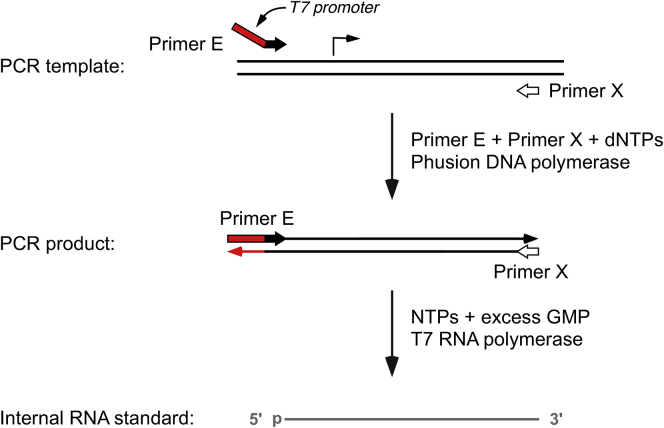
Figure 2.
Synthesis of a cognate internal standard
A DNA template for RNA synthesis is generated by PCR amplification of the gene of interest (parallel black lines) with two transcript-specific primers: primer E (broad red and black arrow), which contains a T7 promoter (red segment) and anneals 10–50 nucleotides upstream of the transcription initiation site (bent black arrow) of the cellular transcript under investigation, and primer X (broad white arrow), which anneals >100 nt downstream. The 10–50 nt extension added by primer E need not match the upstream region of the gene encoding the cellular transcript under investigation as long as the primer can anneal near the initiation site of that gene. In vitro transcription of the PCR product by T7 RNA polymerase in the presence of a 30-fold molar excess of GMP over GTP generates a cognate internal standard (gray line) bearing a monophosphate (p) at its 5′ end.
Final analysis
Timing: 21 h
Using RT-PCR to measure the percentage of a cellular transcript that is monophosphorylated requires comparison to a cognate internal standard that itself is monophosphorylated and can be reverse transcribed and replicated with the same set of primers. To be informative, the internal standard must be added at a concentration that is comparable to that of the transcript(s) under investigation. This concentration is determined empirically by serially diluting the internal standard and spiking equal amounts (0.001–10 ng) into samples of cellular RNA (7.5 μg) that have or have not been treated with RppH. The resulting pairs of mixtures are then ligated to the chimeric oligonucleotide, reverse transcribed, PCR amplified, and examined by electrophoresis as described for the preliminary analysis.
RppH treatment
Perform as described for the preliminary analysis.
Ligation of a chimeric oligonucleotide
Perform as described for the preliminary analysis (Figure 1), but include the monophosphorylated internal standard.
| Nucleic acid mixture | |
|---|---|
| Reagent | Amount for one reaction (μL) |
| Cellular RNA (0.75 μg/μL) (± RppH treatment) | 10.0 |
| Monophosphorylated RNA standard | 5.0 |
| Chimeric oligonucleotide (1 μg/μL) | 1.0 |
| Subtotal | 16.0 |
Note: An additional nucleic acid mixture should contain only the monophosphorylated internal standard and the chimeric oligonucleotide, with 7.5 μg of tRNA, poly(A), or total RNA from another species substituted for the cellular RNA listed above. Parallel analysis of this additional mixture enables the bands on the final polyacrylamide gel to be identified correctly.
-
31.
Incubate the nucleic acid mixture at 65°C for 5 min and then transfer it to ice for 1 min.
-
32.
Add 9 μL of ligase master mix to each tube.
| Ligase master mix | |
|---|---|
| Reagent | Amount for one reaction (μL) |
| Water | 4.25 |
| 10× T4 RNA ligase buffer | 2.5 |
| ATP (100 mM) | 0.25 |
| RNasin (40 U/μL) | 0.5 |
| T4 RNA ligase 1 (10 U/μL) | 1.5 |
| Subtotal | 9.0 |
| Reagent | Amount for one reaction (μL) |
|---|---|
| Nucleic acid mixture | 16.0 |
| Ligase master mix | 9.0 |
| Total | 25.0 |
-
33.
Mix thoroughly and incubate at 37°C for 2 h.
-
34.
Add 125 μL of 3 mM EDTA (pH 8.0), extract with phenol/chloroform (pH 4.3), and ethanol precipitate as described in step 3. Dissolve the RNA pellets in 11.5 μL of sterile water.
Reverse transcription, amplification, and electrophoresis
Perform as described for the preliminary analysis (Figure 1). Quantify as described below.
Quantitative analysis of PABLO-QA data
The final stage of PABLO-QA concludes with quantifying band intensities and calculating the percentage of 5′ ends that are monophosphorylated. The two gel lanes (±RppH, spiked with identical amounts of the internal standard) in which the intensity of the DNA band representing the monophosphorylated internal standard is most similar to that for the transcript(s) of interest are scanned and quantified with ImageJ or commercial software to determine the relative intensities of those bands. Because ethidium bromide staining can result in substantial fluorescence between DNA bands, background correction is essential. Finally, the percentage of each cellular transcript that is monophosphorylated is calculated by comparing the relative band intensities with or without prior treatment of the cellular RNA with excess RppH, as follows:
The experiments (±RppH) with the optimal concentration of the internal standard should be performed in triplicate to allow mean values and standard deviations to be calculated.
Expected outcomes
PABLO-QA is a sensitive method for accurately measuring the percentage of any particular RNA 5′ end that is monophosphorylated, even when that percentage is small. It is particularly useful for examining the phosphorylation state of RNAs whose cellular concentration is low or that have multiple closely spaced 5′ termini. It can be used to differentiate RNA processing sites from transcription initiation sites, as the former will generally be 100% monophosphorylated whereas the latter will typically be more heterogeneous, potentially comprising a mixture of triphosphorylated, diphosphoryated, monophosphorylated, and/or capped 5′ ends caught at various stages of maturation or deprotection. For a 5′ terminus generated by transcription initiation, the percentage that is monophosphorylated reflects the relative rates of formation and decay of the monophosphorylated intermediate. In combination with genetic mutations, this information can be used to identify the RNA features and proteins that govern these processes.
We have used PABLO-QA to examine the phosphorylation state of the three principal 5′ ends of sugE mRNA in Legionella pneumophila (Richards and Belasco, 2021). A preliminary analysis of the kind described above identified the location of these termini and led us to design a cognate internal standard whose 5′ end preceded that of the longest sugE transcript by 19 nt (Figure 3). After spiking this fully monophosphorylated standard into total RNA from Legionella, a final PABLO-QA analysis was performed to determine the percentage of each sugE 5′ end that was monophosphorylated in vivo (Figure 4, Table 1). In addition, the quantitative reliability of PABLO-QA was verified by analyzing a set of in vitro transcribed sugE RNA mixtures in which the 5′ phosphorylation state of the RNA was known in advance (Figure 5).
Figure 3.
Preliminary analysis of Legionella pneumophila sugE 5′ ends by PABLO-QA
Total RNA extracted from L. pneumophila by the hot phenol method (Richards and Belasco, 2021) was analyzed by PABLO-QA with sugE-specific primers in the absence of an internal standard, with or without prior treatment of the RNA with excess RppH. The sugE-specific primers X (AATCAGTGTGGTGGCTGTAA) and Y (TGTCTTAACCGGGTACGG) used for this analysis annealed 0.18 kb or 15–22 nt, respectively, downstream of the heterogeneous 5′ ends of sugE mRNA. Above the bands representing the sugE 5′ ends is a clear zone where no bands are visible. For comparison, a cognate internal standard whose principal PABLO-QA product migrates in the clear zone was analyzed in parallel. This internal standard comprised sugE mRNA bearing a 5′-terminal 19-nt extension derived from the sugE promoter region. It was synthesized by in vitro transcription of a DNA template prepared by PCR amplification of L. pneumophila DNA with sugE-specific primers, one of which contained a T7 promoter (underlined) followed by 22 nt of the sugE promoter and transcription unit (lowercase): CCAAAAGAATTCCAAATTAATACGACTCACTATTagtgatatgctataaaataatc.
Figure 4.
Final analysis of Legionella pneumophila sugE 5′ ends by PABLO-QA
(Left) Total RNA extracted from L. pneumophila was analyzed by PABLO-QA with sugE-specific primers X and Y in the presence of the monophosphorylated sugE internal standard described in Figure 3, with or without prior treatment of the cellular RNA with excess RppH. (Right) The intensities of four bands representing the internal standard and three distinct sugE 5′ termini (P1, P2, and P3) were quantified and used to calculate the percentage of each sugE 5′ end that was monophosphorylated. Each value is the average of three biological replicates. Error bars correspond to standard deviations. Modified from Figure 5D of Richards and Belasco (2021).
Table 1.
Example: calculating the percentage of monophosphorylated 5′ ends from PABLO-QA band intensities
| Transcript | Band intensity (raw data) |
Normalized band intensity (transcript/standard) |
% MonoP a |
||
|---|---|---|---|---|---|
| – RppH | + RppH | – RppH | + RppH | – RppH/+ RppHb | |
| sugE standard | 2,986,575 | 1,085,950 | 1.000 | 1.000 | |
| sugE P1 | 307,475 | 2,337,900 | 0.103 | 2.153 | 5 |
| sugE P2 | 2,728,350 | 2,279,975 | 0.914 | 2.100 | 44 |
| sugE P3 | 2,106,750 | 2,310,375 | 0.705 | 2.128 | 33 |
Calculated percentage of each 5′ end that is monophosphorylated.
Calculated from the ratio of normalized band intensities.
Figure 5.
Quantitative accuracy of PABLO-QA
(Top) Monophosphorylated (MonoP) and triphosphorylated (TriP) forms of sugE model RNA synthesized by in vitro transcription were combined at six different molar ratios and spiked with equal amounts of the fully monophosphorylated sugE internal standard described in Figure 3. The mixtures were then analyzed by PABLO-QA with sugE-specific primers. (Bottom) The percentage of sugE model RNA that was monophosphorylated was calculated from the relative band intensities in each lane and compared to the actual percentage of monophosphorylated sugE model RNA in the original mixture. In these calculations, the ratio of band intensities in the lane representing fully monophosphorylated sugE model RNA (100% monoP) served as a surrogate for the ratio obtained with an RppH-treated sample. Each value is the average of three independent measurements. Error bars (some too small to see) correspond to standard deviations. The line represents an ideal experimental outcome. Reproduced from Figure 5C of Richards and Belasco (2021).
Limitations
Transcripts whose abundance is exceptionally low
Even though the use of PCR amplification makes PABLO-QA very sensitive, some transcripts may be so scarce that PCR cannot amplify their signal above that of non-specific amplification products.
Stable secondary structure that masks the 5′ end
In principle, sequestration of an RNA 5′ end in thermodynamically stable base pairing might impair its reactivity with RppH and/or T4 RNA ligase 1. Nevertheless, we have successfully used PABLO-QA to determine the phosphorylation state of a transcript with only one unpaired nucleotide at the 5′ end.
Troubleshooting
Problem 1
Low yield of cellular RNA.
Potential solution
Thanks to PCR amplification, the mRNA of interest may be sufficiently abundant to allow less cellular RNA to be used in each reaction.
Problem 2
Structural obstacle to reverse transcription.
Potential solution
Additional PCR cycles may compensate for a low yield of full-length reverse transcription products caused by premature termination.
Problem 3
Non-specific amplification products.
Potential solution
This problem can be solved by redesigning primers X and Y or by increasing the PCR annealing temperature.
Problem 4
The DNA sequencing electropherogram obtained during 5′ end mapping contains overlaid peaks due to a mixed population of PCR products.
Potential solution
Overlaid DNA sequences indicate inadequate separation of the PCR products generated by amplification with primers B and Y. The electrophoretic resolution of these PCR products can be improved by changing the percentage of polyacrylamide or by allowing the PCR products to migrate further.
Problem 5
High background fluorescence on polyacrylamide gels stained with ethidium bromide.
Potential solution
Background fluorescence can be reduced by destaining the gel in 1× TBE for 10 min. If this does not resolve the problem, then use of a fluorescently labeled primer in the second round of PCR would obviate the need for ethidium staining.
Resource availability
Lead contact
Inquiries about the protocol should be addressed to the lead contact, Joel Belasco (joel.belasco@med.nyu.edu).
Materials availability
The materials and reagents needed for this protocol are commercially available.
Acknowledgments
This research was supported by a grant to J.G.B. from the National Institutes of Health (R01GM123124).
Author contributions
J.G.B. conceived of the method, J.R. and J.G.B. designed the procedure. J.R. validated the procedure empirically, and J.R. and J.G.B. wrote the protocol.
Declaration of interests
The authors declare no competing interests.
Contributor Information
Jamie Richards, Email: jamie.richards@med.nyu.edu.
Joel G. Belasco, Email: joel.belasco@med.nyu.edu.
Data and code availability
All of the pertinent data are presented in Table 1 and Figures 3, 4, and 5. No computer code was generated in the course of this study.
References
- Bandyra K.J., Said N., Pfeiffer V., Gorna M.W., Vogel J., Luisi B.F. The seed region of a small RNA drives the controlled destruction of the target mRNA by the endoribonuclease RNase E. Mol. Cell. 2012;47:943–953. doi: 10.1016/j.molcel.2012.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bischler T., Hsieh P.K., Resch M., Liu Q., Tan H.S., Foley P.L., Hartleib A., Sharma C.M., Belasco J.G. Identification of the RNA pyrophosphohydrolase RppH of Helicobacter pylori and global analysis of its RNA targets. J. Biol. Chem. 2017;292:1934–1950. doi: 10.1074/jbc.M116.761171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cahová H., Winz M.L., Höfer K., Nübel G., Jäschke A. NAD captureSeq indicates NAD as a bacterial cap for a subset of regulatory RNAs. Nature. 2015;519:374–377. doi: 10.1038/nature14020. [DOI] [PubMed] [Google Scholar]
- Celesnik H., Deana A., Belasco J.G. Initiation of RNA decay in Escherichia coli by 5' pyrophosphate removal. Mol. Cell. 2007;27:79–90. doi: 10.1016/j.molcel.2007.05.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celesnik H., Deana A., Belasco J.G. PABLO analysis of RNA: 5'-phosphorylation state and 5'-end mapping. Methods Enzymol. 2008;447:83–98. doi: 10.1016/S0076-6879(08)02205-2. [DOI] [PubMed] [Google Scholar]
- Deana A., Celesnik H., Belasco J.G. The bacterial enzyme RppH triggers messenger RNA degradation by 5' pyrophosphate removal. Nature. 2008;451:355–358. doi: 10.1038/nature06475. [DOI] [PubMed] [Google Scholar]
- German M.A., Luo S., Schroth G., Meyers B.C., Green P.J. Construction of Parallel Analysis of RNA Ends (PARE) libraries for the study of cleaved miRNA targets and the RNA degradome. Nat. Protoc. 2009;4:356–362. doi: 10.1038/nprot.2009.8. [DOI] [PubMed] [Google Scholar]
- Luciano D.J., Belasco J.G. Analysis of RNA 5′ ends: phosphate enumeration and cap characterization. Methods. 2019;155:3–9. doi: 10.1016/j.ymeth.2018.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luciano D.J., Vasilyev N., Richards J., Serganov A., Belasco J.G. A novel RNA phosphorylation state enables 5′ end-dependent degradation in Escherichia coli. Mol. Cell. 2017;67:44–54. doi: 10.1016/j.molcel.2017.05.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muhlrad D., Decker C.J., Parker R. Deadenylation of the unstable mRNA encoded by the yeast MFA2 gene leads to decapping followed by 5'→3' digestion of the transcript. Genes Dev. 1994;8:855–866. doi: 10.1101/gad.8.7.855. [DOI] [PubMed] [Google Scholar]
- Richards J., Belasco J.G. Widespread protection of RNA cleavage sites by a riboswitch aptamer that folds as a compact obstacle to scanning by RNase E. Mol. Cell. 2021;81:127–138. doi: 10.1016/j.molcel.2020.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards J., Liu Q., Pellegrini O., Celesnik H., Yao S., Bechhofer D.H., Condon C., Belasco J.G. An RNA pyrophosphohydrolase triggers 5′-exonucleolytic degradation of mRNA in Bacillus subtilis. Mol. Cell. 2011;43:940–949. doi: 10.1016/j.molcel.2011.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All of the pertinent data are presented in Table 1 and Figures 3, 4, and 5. No computer code was generated in the course of this study.