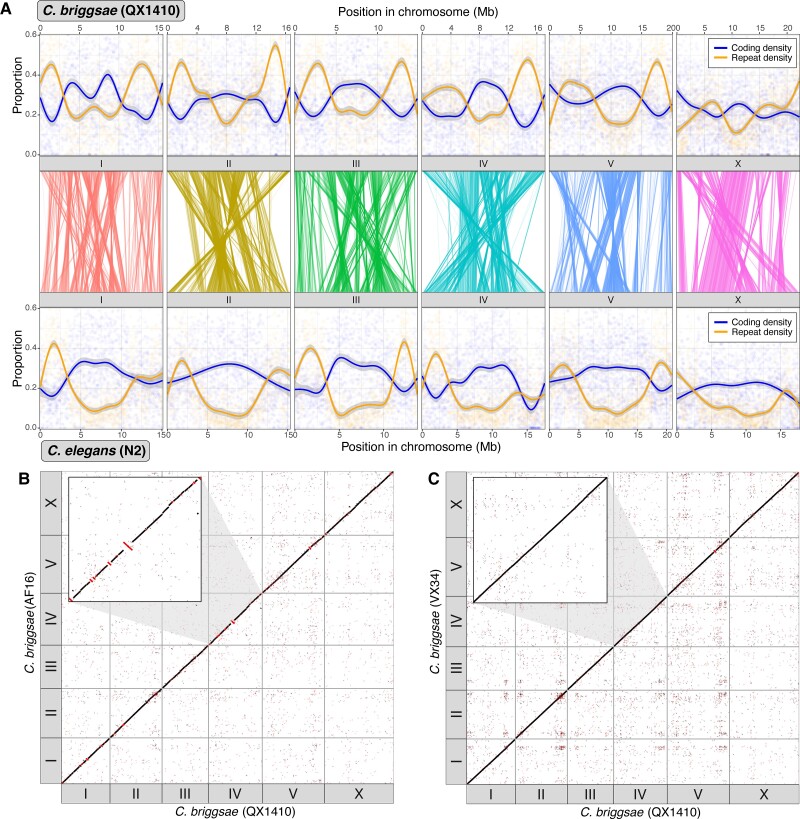
Fig. 1.
High-quality reference genomes for two C. briggsae strains. (A) Comparison between the C. briggsae QX1410 and C. elegans N2 reference genomes. Repeat density and protein-coding gene density per 10 kb windows are shown. Repeats were identified de novo using RepeatModeler2. Solid lines represent LOESS smoothing functions fitted to the data. Relative positions of 10,387 one-to-one orthologs are shown as lines joining the two density plots. (B) Whole-genome alignment of AF16 to the QX1410 reference genome generated using nucmer. Alignments shorter than 1 kbp are not shown. Alignments in the reverse orientation are highlighted in red. Inset: chromosome IV showing multiple regions between AF16 and QX1410 that are in different orientations. (C) Whole-genome alignment of VX34 to the QX1410 reference genome generated using nucmer. Alignments shorter than 1 kbp are not shown. Alignments in the reverse orientation are highlighted in red. Inset: the same chromosome IV region as in (B) showing a largely collinear alignment.