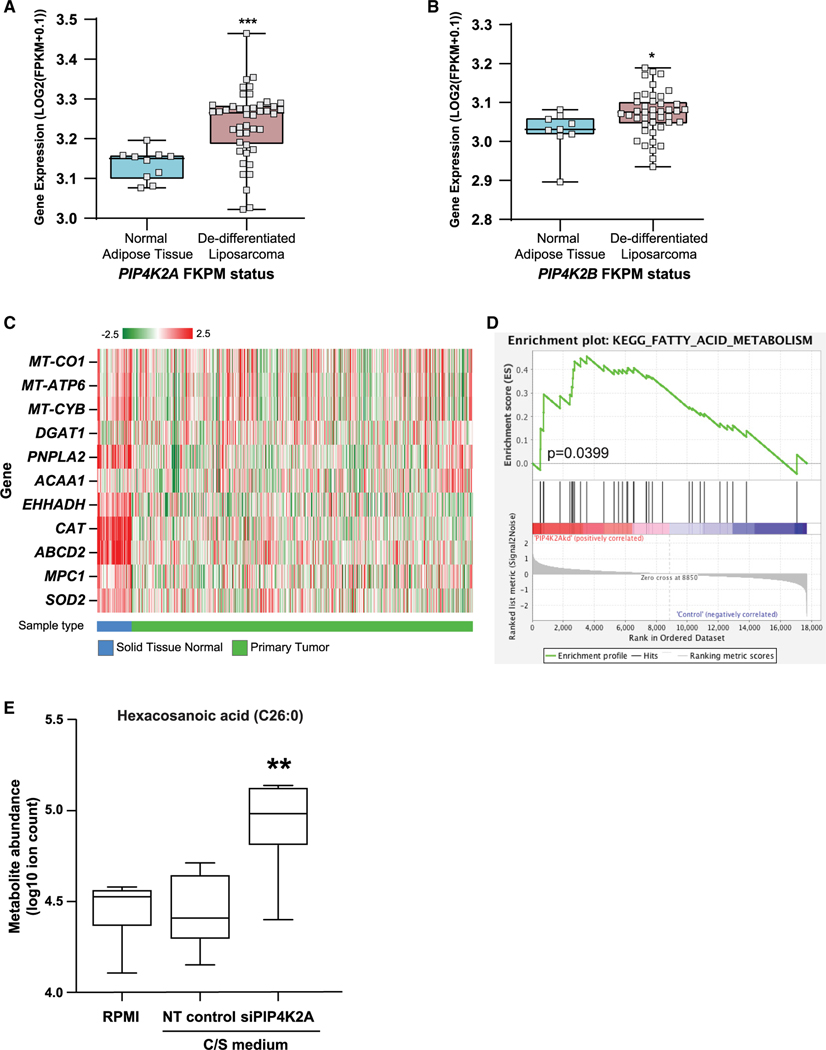
Figure 6. Data from cancer samples indicate a role for PI5P4Ks in regulating FA metabolism.
(A and B) Gene expression of (A) PIP4K2A and (B) PIP4K2B in de-differentiated liposarcoma samples as compared with normal adipose tissue. Statistical significance was determined by comparison of the gene between two groups (Barretina et al., 2010).
(C) Heatmap showing gene expression for TCGA-BRCA normal tissue versus primary tumor samples, exemplifying a number of select genes important for the(peroxisome-mitochondrial function). Red to green color represents high to low log2(FPKM+0.1) expression.
(D) RNA-seq of siPIP4K2A human prostate cancer cells, LNCaP, show an enrichment of FA metabolism compared with the non-targeting (NT) control.
(E) LNCaP cells transfected with siRNA targeting PIP4K2A, cultured in charcoal stripped (C/S) medium, have enriched hexacosanoic acid (C26:0) by mass spectrometry. Transfection of NT siRNAs used as control. Statistical analysis: an unpaired two-tailed t test for used for (A and B) and a paired two-tailed t test was used for (E).