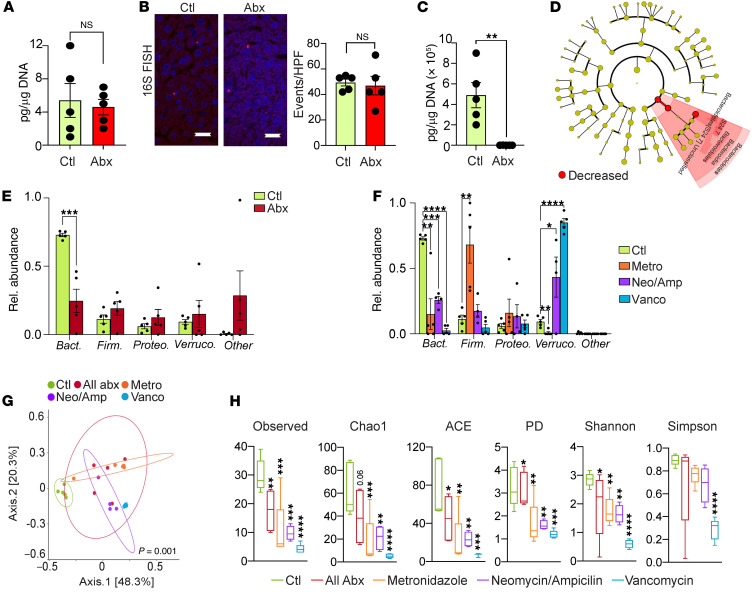
Figure 5. Antibiotic administration selectively modulates the bacterial composition of the liver microbiome.
(A) Hepatic bacterial DNA content was comparatively analyzed by qPCR in cohorts of 6-week-old female mice treated with broad-spectrum antibiotics or vehicle. n = 5/group. (B) The intrahepatic microbiome was compared in the liver of mice treated with broad-spectrum antibiotics (Abx) or vehicle (Ctl) by 16S FISH. Representative images and quantitative results are shown. n = 5/group. Scale bars: 20 μm. (C) Gut bacterial DNA content was comparatively analyzed by qPCR in cohorts of 6-week-old female mice treated with broad-spectrum antibiotics or vehicle. n = 5/group. **P < 0.01. (D) Cladogram showing changes in abundance of microbes across the entire taxonomic hierarchy in the liver of mice treated with broad-spectrum antibiotics. (E) Taxonomic composition of microbiota assigned to phylum level in the liver of mice treated with broad-spectrum antibiotics or vehicle determined by 16S rRNA-Seq. n = 5/group. ***P < 0.001. (F) Taxonomic composition of microbiota assigned to phylum level in the liver of mice treated with selective antibiotics or vehicle determined by 16S rRNA-Seq. n = 5/group. *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001. (G) Weighted PCoA plots of hepatic microbial communities based on Bray-Curtis dissimilarity matrix. Each symbol represents a sample from the liver microbiome of a mouse treated with broad-spectrum or selective antibiotics or vehicle. Clusters were determined by pairwise PERMANOVA. The x and y axes indicate percentage of variation, and ellipses indicate 95% CI. (H) The liver microbiomes in mice treated with broad-spectrum or selective antibiotics were analyzed for α-diversity measures compared with mice treated with vehicle. n = 5/group. *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001.