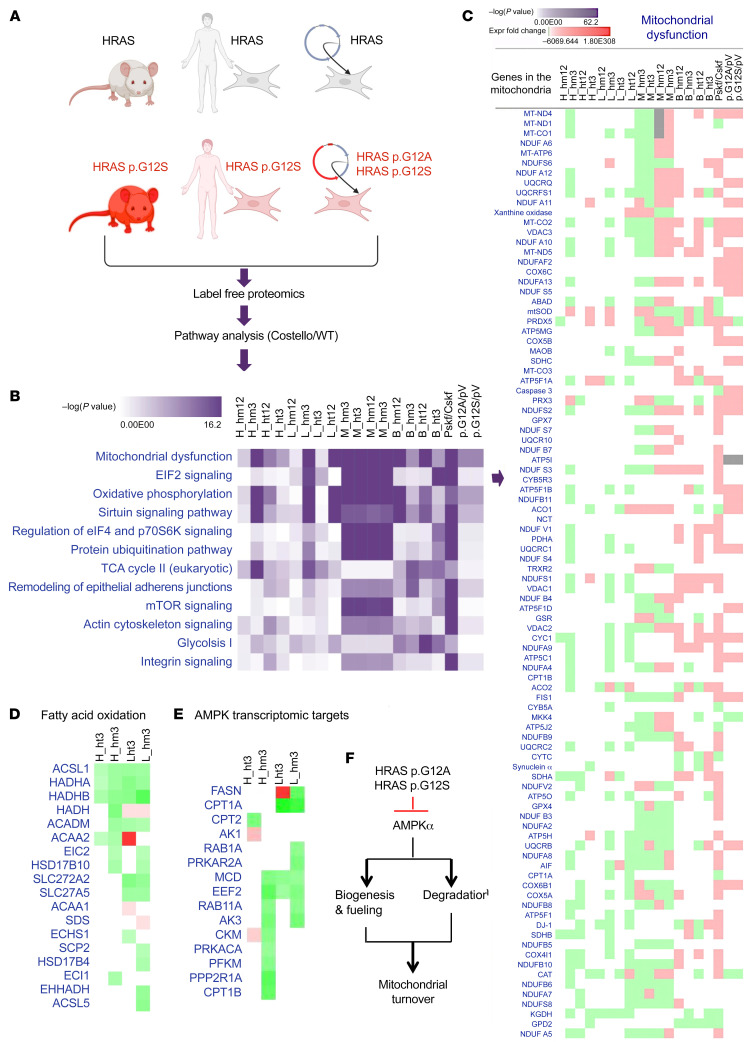
Figure 3. Mitochondrial proteostasis is compromised in Costello syndrome.
(A) Description of the 3 types of Costello syndrome (CS) models used in the label-free proteomic analysis. A differential proteome was obtained between WT and CS models. Ingenuity Pathway Analysis (Qiagen; version 01-07) was used to perform a comparative analysis of the data. n = 3 was used for each sample. (B) Top canonical pathways altered in the CS biological models [ranked by –log(P value)]. (C) Detail of the changes in the “mitochondrial dysfunction” category (expression fold change; green, downregulation; red, upregulation). (D and E) Changes in the fatty acid oxidation and AMPK signaling (indirect transcriptomic targets) proteins detected in the hearts and livers of 3-week-old CS mice (heterozygous or homozygous HRAS p.G12S) compared with WT mice. (F) Working hypothesis on the effect of mutant HRAS p.G12A and HRAS p.G12S on mitochondrial biogenesis and degradation via inhibition of AMPK signaling. H, heart; B, brain; L, liver; M, skeletal muscle; ht, heterozygous; hm, homozygous; 3, 3-week-old mice; 12, 12-week-old mice; Pskf, patient skin fibroblasts; Cskf, control skin fibroblasts; p.G12A, human skin fibroblasts transduced with HRAS p.G12A–expressing lentiviral plasmid; p.G12S, human skin fibroblasts transduced with HRAS p.G12S–expressing lentiviral plasmid; pV, human skin fibroblasts transduced with empty lentiviral plasmid.