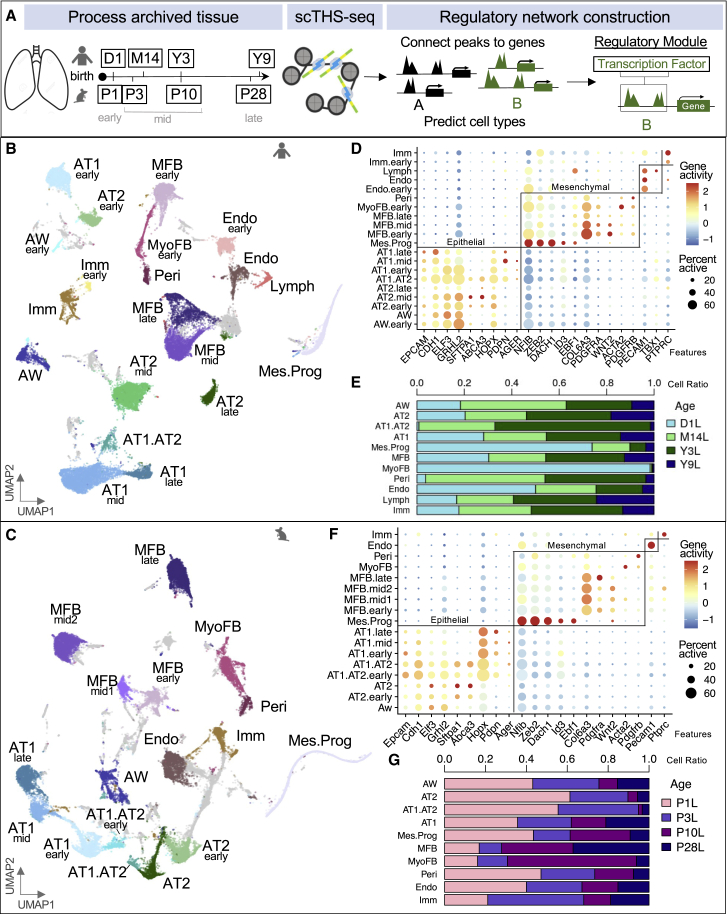
Figure 1.
Lung cell types defined by scTHS-seq chromatin profiles
(A) Overview of workflow with single-cell isolation from archived lung tissue, application of scTHS-seq, and downstream de-novo cell-type identification and construction of regulatory modules. Human and mouse icons indicate corresponding species. For age alignment, a post-weaning, pre-pubertal human to mouse estimation of 1 human year equals 3.65 mouse days was used after postnatal day 1.15
(B and C) UMAP of human (40,427 cells) and mouse (41,476 cells) cell types predicted using peaks in promoter regions of marker genes and Cicero gene activity scores. Based on the majority of cells from different donor ages in each cluster, cell types were further annotated as early (day 1), mid (2 middle time points), or late (oldest time point). Each dot represents a cell and gray clusters represent un-classified populations.
(D and F) Dot plots of log-normalized gene activity scores for select genes in each cell type. The size of each dot corresponds to the percent of cells in that cell type with non-zero gene activity while the color corresponds to the average gene activity.
(E and G) Cell-type contribution from human and mouse lung developmental time point captured as a ratio of total cells.
See also Figures S1 and S2 and Tables S1 and S2.